Dynamic Spectrum and Imaging: Difference between revisions
Jump to navigation
Jump to search
| Line 27: | Line 27: | ||
domedian =True | domedian =True | ||
## this step generates a dynamic spectrum and saves it to IDB20170821201020-203020.12s.slfcaled.ms.dspec.npz | ## this step generates a dynamic spectrum and | ||
## saves it to IDB20170821201020-203020.12s.slfcaled.ms.dspec.npz | |||
ds.get_dspec(vis=msfile, specfile=specfile, bl=bl, spw=spw, domedian=domedian) | ds.get_dspec(vis=msfile, specfile=specfile, bl=bl, spw=spw, domedian=domedian) | ||
Revision as of 14:59, 15 May 2019
Get Dynamic Sepctrum with SunCASA
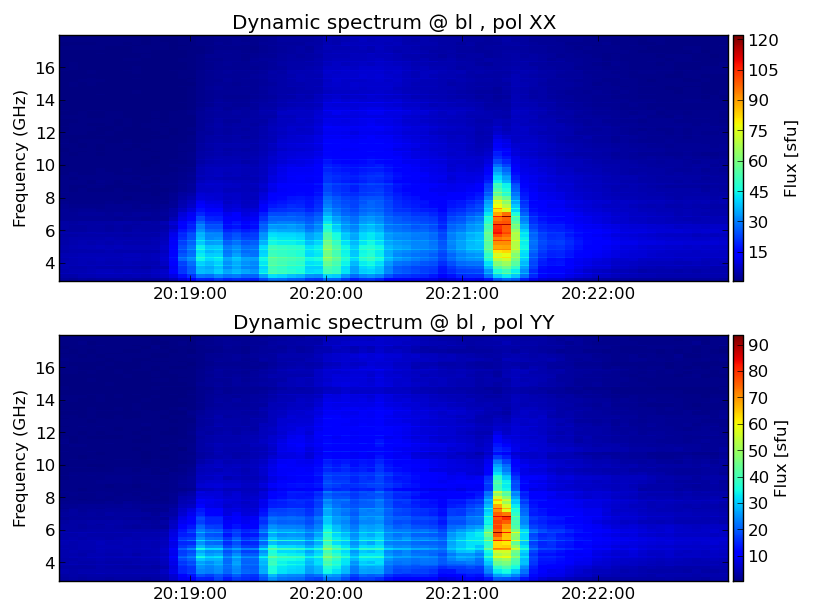
Within SunCASA, you are using IPython to interact with the system. This does not mean extensive python experience is necessary. Basic Python interactions are straightforward, e.g., assigning parameters, importing modules, running functions. The first module we introduce is dspec. This module allows you to generate a dynamic spectrum from an MS file, and visualize it. You can select a subset of data by specifying a time range, spectral windows/channels, antenna baseline. The selection syntax follows the CASA convention. More information may be found in the CASA guide of time range, spectral windows/channels, antenna baseline selection pages.
from suncasa.utils import dspec as ds # define the visbility data file msfile = 'IDB20170821201020-203020.12s.slfcaled.ms' ## define the output filename of the dynamic spectrum specfile = msfile + '.dspec.npz' ## antenna selection ## leave it blank for selecting all baselines bl = '' ## Spectral Windows and Channels selection leave it blank for selecting all spectral windows spw = '' ## time range selection ## leave it blank for selecting the entire time interval timeran = '' ## select baselines with a median length (0.2~0.8km) domedian =True ## this step generates a dynamic spectrum and ## saves it to IDB20170821201020-203020.12s.slfcaled.ms.dspec.npz ds.get_dspec(vis=msfile, specfile=specfile, bl=bl, spw=spw, domedian=domedian) ## this plot the dynamic spectrum. ds.plt_dspec(specfile, pol='XXYY')
Imaging with SunCASA
cd to your working directory where the measurement sets file is located.
from suncasa.utils import qlookplot as ql msfile = 'IDB20170821201020-203020.12s.slfcaled.ms' vis = msfile timerange = '20:21:10~20:21:30' ## time range selection spw = '3.4~6.0GHz' ## Spectral Windows and Channels selection stokes = 'XXYY' ## polarizations selection ql.qlookplot(vis, timerange=timerange, spw=spw, stokes=stokes)