EOVSA Data Analysis Tutorial: Difference between revisions
No edit summary |
|||
| (187 intermediate revisions by 4 users not shown) | |||
| Line 12: | Line 12: | ||
=Calibrating EOVSA data= | =Calibrating EOVSA data= | ||
*<b>Calibration</b>: | *<b>Calibration</b>: Calibrating EOVSA data is an involved process. If you are interested the [[Practical Calibration Tutorial]] describes the procedure. We are currently working on a semi-automatic pipeline for calibrating the visibility data. At this moment, however, please contact the EOVSA team if you wish to have calibrated visibility data for a specific event. In the near future, we envision the pipeline-calibrated visibility data to be available for download on a regular basis. | ||
*<b>Self-calibration</b>: For improving the quality of the imaging, self-calibration is usually needed. This is still a work-in-progress, but we have had some successful practice in self-calibrating some flare events. Please refer to [[Self-Calibrating Flare Data]] for examples and detailed discussions on our current practice for self-calibrating EOVSA flare data. | *<b>Self-calibration</b>: For improving the quality of the imaging, self-calibration is usually needed. This is still a work-in-progress, but we have had some successful practice in self-calibrating some flare events. Please refer to [[Self-Calibrating Flare Data]] [[Tohban_EOVSA_Imaging_Tutorial_A-Z]] for examples and detailed discussions on our current practice for self-calibrating EOVSA flare data. | ||
=Analyzing EOVSA data= | =Analyzing EOVSA data= | ||
==Software== | ==Software== | ||
We have developed two packages for EOVSA data processing and analysis: | We have developed two packages for EOVSA data processing and analysis: | ||
* [https://github.com/ | * [https://github.com/suncasa/suncasa SunCASA] A wrapper around [https://casa.nrao.edu/ CASA (the Common Astronomy Software Applications package)] for synthesis imaging and visualizing solar spectral imaging data. CASA is one of the leading software tool for "supporting the data post-processing needs of the next generation of radio astronomical telescopes such as ALMA and VLA", an international effort led by the [https://public.nrao.edu/ National Radio Astronomy Observatory]. The current version of CASA uses Python (2.7) interface. More information about CASA can be found on [https://casa.nrao.edu/ NRAO's CASA website ]. Note, CASA is available ONLY on UNIX-BASED PLATFORMS (and therefore, so is SunCASA). | ||
* [https://github.com/Gelu-Nita/GSFIT GSFIT] A IDL-widget(GUI)-based spectral fitting package called gsfit, which provides a user-friendly display of EOVSA image cubes and an interface to fast fitting codes (via platform-dependent shared-object libraries). | * [https://github.com/Gelu-Nita/GSFIT GSFIT] A IDL-widget(GUI)-based spectral fitting package called gsfit, which provides a user-friendly display of EOVSA image cubes and an interface to fast fitting codes (via platform-dependent shared-object libraries). | ||
For this tutorial, there are two approaches in accessing our software packages: | |||
* 1. Through our '''Amazon Cloud server'''. See [[#Using Software on AWS server|Section 3.1.1]] for detailed instructions. | |||
* 2. Install on '''your own machine'''. See [[#SunCASA Installation|Section 3.1.2]] (SunCASA) and [[#GSFIT Installation|Section 3.1.3]] (GSFIT) for instructions. | |||
===Using Software on Amazon Cloud Server=== | |||
We use an [https://aws.amazon.com/lightsail/ Amazon AWS Lightsail] cloud server for lightweight data processing and testing purposes. The server has 2 CPUs, 8 GB RAM, and 160 GB SSD storage. It runs CentOS 7 (1901-01) Linux. | |||
''NOTE: the guest account is <b>ONLY INTENDED FOR THE EOVSA TUTORIAL, WHICH WILL BE DISABLED SHORTLY AFTER THE RHESSI WORKSHOP </b> (you will be notified prior to that). '' If you wish to continue using the server for testing purposes after the tutorial, please contact [mailto:bin.chen@njit.edu Bin Chen] to set up a personal account. | |||
'' | ''DISCLAIMER: We are NOT RESPONSIBLE for any loss, damage, or leak of your personal data on the server.'' | ||
* Obtain SSH Key from [mailto:bin.chen@njit.edu Bin Chen] | * Obtain SSH Key to the guest account from [mailto:bin.chen@njit.edu Bin Chen] | ||
* Put it under a secure location on your own machine. | * Put it under a secure location on your own machine. | ||
* Follow remaining directions depending on your client machine. | * Follow remaining directions depending on your client machine. | ||
====Connecting via | ====Connecting via command line==== | ||
* Edit the permission of the SSH key and the directory you choose to place it. Here I use ~/.ssh as an example (create it first by ''mkdir ~/.ssh'' if it does not exist.) | |||
<pre> | <pre> | ||
chmod 700 ~/.ssh | chmod 700 ~/.ssh | ||
chmod 400 ~/.ssh/guest-virgo.pem | chmod 400 ~/.ssh/guest-virgo.pem | ||
</pre> | </pre> | ||
* Log on to | * Log on to AWS server (password-less) | ||
<pre> | <pre> | ||
ssh -X -i ~/.ssh/guest-virgo.pem guest@virgo.arcs.az.njit.edu | ssh -X -i ~/.ssh/guest-virgo.pem guest@virgo.arcs.az.njit.edu | ||
</pre> | </pre> | ||
Now you are connected to '''virgo'''. | Now you are connected to '''virgo'''. | ||
====Connecting via Windows (MobaXterm)==== | ====Connecting via Windows (MobaXterm)==== | ||
Recommend to use "Documents\MobaXterm\home\.ssh", which should exist if you have already installed the free MobaXterm[https://mobaxterm.mobatek.net/]. | Recommend to use "Documents\MobaXterm\home\.ssh", which should exist if you have already installed the free MobaXterm[https://mobaxterm.mobatek.net/]. | ||
* Create new session, click SSH, enter virgo.arcs.az.njit.edu for Remote host, guest for username. | * Create new session, click SSH, enter virgo.arcs.az.njit.edu for Remote host, guest for username. | ||
* On advanced SSH settings tab, click Use private key, navigate to and select file | * On advanced SSH settings tab, click Use private key, navigate to and select file guest_virgo.pem | ||
* Close setup window and click the new | * Close setup window and click the new session's icon, which will log you in. | ||
====Startup SunCASA and sswIDL==== | ====Startup SunCASA and sswIDL==== | ||
Once | Once you are connected to '''virgo''', you will have access to SunCASA and GSFIT (included in the sswIDL installation). To not interfere with others (who share the same "guest" account), please <b>create your own directory and work under it</b>. For easier identification, please kindly use <b>the initial of your first name and your full last name as the name of your directory</b> (here "bchen" is used as an example). | ||
<pre> | <pre> | ||
[guest@ip-172-26-5-203 ~]$ mkdir bchen | [guest@ip-172-26-5-203 ~]$ mkdir bchen | ||
[guest@ip-172-26-5-203 ~]$ cd bchen | [guest@ip-172-26-5-203 ~]$ cd bchen | ||
</pre> | </pre> | ||
To enter SunCASA | |||
<pre> | <pre> | ||
[guest@ip-172-26-5-203 ~/bchen]$ suncasa | [guest@ip-172-26-5-203 ~/bchen]$ suncasa | ||
</pre> | </pre> | ||
To enter sswIDL | |||
<pre> | <pre> | ||
[guest@ip-172-26-5-203 ~/bchen]$ sswidl | [guest@ip-172-26-5-203 ~/bchen]$ sswidl | ||
| Line 68: | Line 70: | ||
===[[SunCASA Installation]]=== | ===[[SunCASA Installation]]=== | ||
Please | Please [http://www.ovsa.njit.edu/wiki/index.php/SunCASA_Installation follow this link] for details regarding installation of SunCASA on your own machine (only available on Unix-bases OS). This will take you to another page. | ||
===[[GSFIT Installation]]=== | ===[[GSFIT Installation]]=== | ||
Please | Please [http://www.ovsa.njit.edu/wiki/index.php/GSFIT_Installation follow this link] for details regarding installation of GSFIT on your own machine (which supports multiple platforms). This will take you to another page. | ||
== | ==Example Data and Scripts== | ||
In this tutorial, we will demonstrate steps for spectral imaging (self-)calibrated visibility data for a C-class flare on 2017 Aug 21 at ~20:20 UT ([[#Spectral Imaging with SunCASA|Section 3.3]]), followed by a tutorial on visualizing and analyzing the resulting spectral imaging cube ([[#Spectral Fitting with GSFIT|Section 3.4]]). | |||
The visibility data can be accessed by | |||
* Downloading from [https://drive.google.com/open?id=1ZMCY9Y3FTv-m0QFyBBwrvqaSuS5pAc3l this Google Drive link]. | |||
* If you are on our AWS cloud server "virgo" (means for accessing the server are discussed in [[#Using Software on AWS server|Section 3.1.1]]), it is located at /common/data/eovsa_tutorial/IDB20170821201800-202300.4s.slfcaled.ms. | |||
For completeness, the pre-slfcalibration visibility data can be downloaded from [https://drive.google.com/open?id=1ZMCY9Y3FTv-m0QFyBBwrvqaSuS5pAc3l this Google Drive link] (not used by this tutorial). | |||
''To reduce the amount of data for this tutorial, the time resolution has been reduced by a factor of 4, to 4 s per sample. The actual time resolution available is 1 s.'' If you are on virgo, please copy it over to your own working directory that you created earlier under the guest account (e.g., ~/bchen/). | |||
<pre style="background-color:#FCEBD9;"> | <pre style="background-color:#FCEBD9;"> | ||
cd | cd ''your_working_directory'' | ||
cp -r /common/data/eovsa_tutorial/IDB20170821201800-202300.4s.slfcaled.ms ./ | cp -r /common/data/eovsa_tutorial/IDB20170821201800-202300.4s.slfcaled.ms ./ | ||
</pre> | </pre> | ||
EOVSA data is handled in [https://casa.nrao.edu CASA] tables system, known as a Measurement Set (MS). The actual visibility data are stored in a MAIN table that contains a number of rows, each of which is effectively a single timestamp for a single spectral window and a single baseline. Within SunCASA, you | Example scripts for doing the analysis covered in this tutorial are available as [https://github.com/binchensun/eovsa-tutorial/tree/master/rhessi18 this Github repository]. On Virgo, the scripts can be found under /common/data/eovsa_tutorial/. | ||
==Spectral Imaging with SunCASA== | |||
EOVSA data is handled in [https://casa.nrao.edu CASA] tables system, known as a Measurement Set (MS). The actual visibility data are stored in a MAIN table that contains a number of rows, each of which is effectively a single timestamp for a single spectral window and a single baseline. Within SunCASA, in addition to everything already available in CASA, you have access to additional tools that allow you to explore and utilize the new radio dynamic spectroscopic imaging data from EOVSA. We also had success in using SunCASA for analyzing data from the [https://science.nrao.edu/facilities/vla Karl G. Jansky Very Large Array]. The steps are very similar to those described here. | |||
To start SunCASA, use: | To start SunCASA, use: | ||
<pre style="background-color:#FCEBD9;"> | <pre style="background-color:#FCEBD9;"> | ||
suncasa | suncasa | ||
</pre> | </pre> | ||
Within SunCASA, you are under the IPython environment. Everything you know about (I)Python should be applicable here. The installation comes with frequently used packages including Matplotlib, Numpy, SciPy, AstroPy, SunPy. However, it is not very intuitive to add (compatible) Python packages within (Sun)CASA. If you need some specific packages for your analysis, and it does not require direct interaction with (Sun)CASA, we recommend you to use the standard Python environment. On Virgo, we have installed Anaconda 3, which can be accessed by, e.g., | Within SunCASA, you are under the IPython environment. Everything you know about (I)Python should be applicable here. The installation comes with frequently used packages including Matplotlib, Numpy, SciPy, AstroPy, SunPy. However, it is not very intuitive to add (compatible) Python packages within (Sun)CASA. If you choose to do so, you may want to check [http://docs.astropy.org/en/stable/install.html#installing-astropy-into-casa this page] on how to install AstroPy (which is not originally shipped with CASA) within CASA. This method is generally applicable to adding other packages (but not thoroughly tested). If you need some specific packages for your analysis, and it does not require direct interaction with (Sun)CASA, we recommend you to use the standard Python environment. On Virgo, we have installed Anaconda 3, which can be accessed by, e.g., typing "ipython" in a terminal window. | ||
===Cross-Power Dynamic Spectrum=== | ===Cross-Power Dynamic Spectrum=== | ||
The first module we introduce is [https://github.com/ | The first module we introduce is [https://github.com/suncasa/suncasa/blob/master/utils/dspec.py ''dspec'']. This module allows you to generate a cross-power dynamic spectrum from an MS file, and visualize it. You can select a subset of data by specifying a [https://casa.nrao.edu/Release3.3.0/docs/UserMan/UserMansu112.html time range], [https://casaguides.nrao.edu/index.php/Selecting_Spectral_Windows_and_Channels spectral windows/channels], [https://casaguides.nrao.edu/index.php/Antenna/Baseline_Selection_Syntax_with_or_without_Autocorrelations antenna baseline], or [https://casa.nrao.edu/Release3.3.0/docs/UserMan/UserMansu113.html uvrange]. The selection syntax follows the ''CASA'' convention. More information of CASA selection syntax may be found in the above links or the [https://casa.nrao.edu/casadocs/casa-5.4.0/data-selection/data-selection-in-a-measurementset Measurement Selection Syntax]. | ||
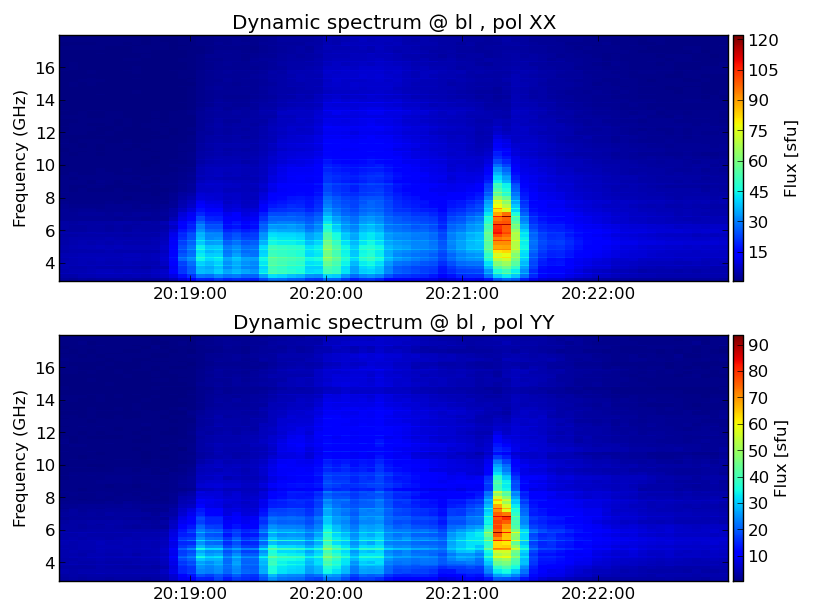
[[file:fig-dspec.png|thumb|right|300px|Figure 1: EOVSA cross power dynamic spectrum at stokes XX and YY]] | [[file:fig-dspec.png|thumb|right|300px|Figure 1: EOVSA cross power dynamic spectrum at stokes XX and YY]] | ||
<pre style="background-color: #FCEBD9;"> | <pre style="background-color: #FCEBD9;"> | ||
| Line 107: | Line 115: | ||
uvrange = '0.15~0.5km' | uvrange = '0.15~0.5km' | ||
## this step generates a dynamic spectrum and saves it to specfile | ## this step generates a dynamic spectrum and saves it to specfile | ||
ds.get_dspec(vis=msfile, specfile=specfile, uvrange=uvrange, domedian=True) | dspec=ds.get_dspec(vis=msfile, specfile=specfile, uvrange=uvrange, domedian=True) | ||
## dspec is a Python dictionary that contains the resulting dynamic spectrum. | |||
## A copy is saved in "specfile" as a numpy npz file. | |||
## Other optional parameters are available for more selection criteria | ## Other optional parameters are available for more selection criteria | ||
## such as frequency range ("spw"), and time range ("timeran") | ## such as frequency range ("spw"), and time range ("timeran") | ||
| Line 115: | Line 125: | ||
pol='XXYY' | pol='XXYY' | ||
## The following command displays the resulting cross-power dynamic spectrum | ## The following command displays the resulting cross-power dynamic spectrum | ||
ds.plt_dspec(specfile, pol=pol) | ds.plt_dspec(specfile, pol=pol) # alternatively you can use the output from the previous step "dspec" as the input | ||
</pre> | </pre> | ||
Now you should have a popup window showing the dynamic spectrum. Hover your mouse over the dynamic spectrum, you can read the time and frequency information at the bottom of the window. | Now you should have a popup window showing the dynamic spectrum. Hover your mouse over the dynamic spectrum, you can read the time and frequency information at the bottom of the window. | ||
===Quick-Look Imaging=== | ===Quick-Look Imaging=== | ||
Imaging EOVSA data involves image cleaning, as well as solar coordinate transformation and image registration. We bundled a number of these steps ino a module named [https://github.com/ | Imaging EOVSA data involves image cleaning, as well as solar coordinate transformation and image registration. We bundled a number of these steps ino a module named [https://github.com/suncasa/suncasa/blob/master/utils/qlookplot.py ''qlookplot''], allowing users to generate an observing summary plot showing the cross power dynamic spectrum, GOES light curves and EOVSA quick-look images. Now let us start with making a summary plot of EOVSA image at the spectral window 5 (5.4 GHz). | ||
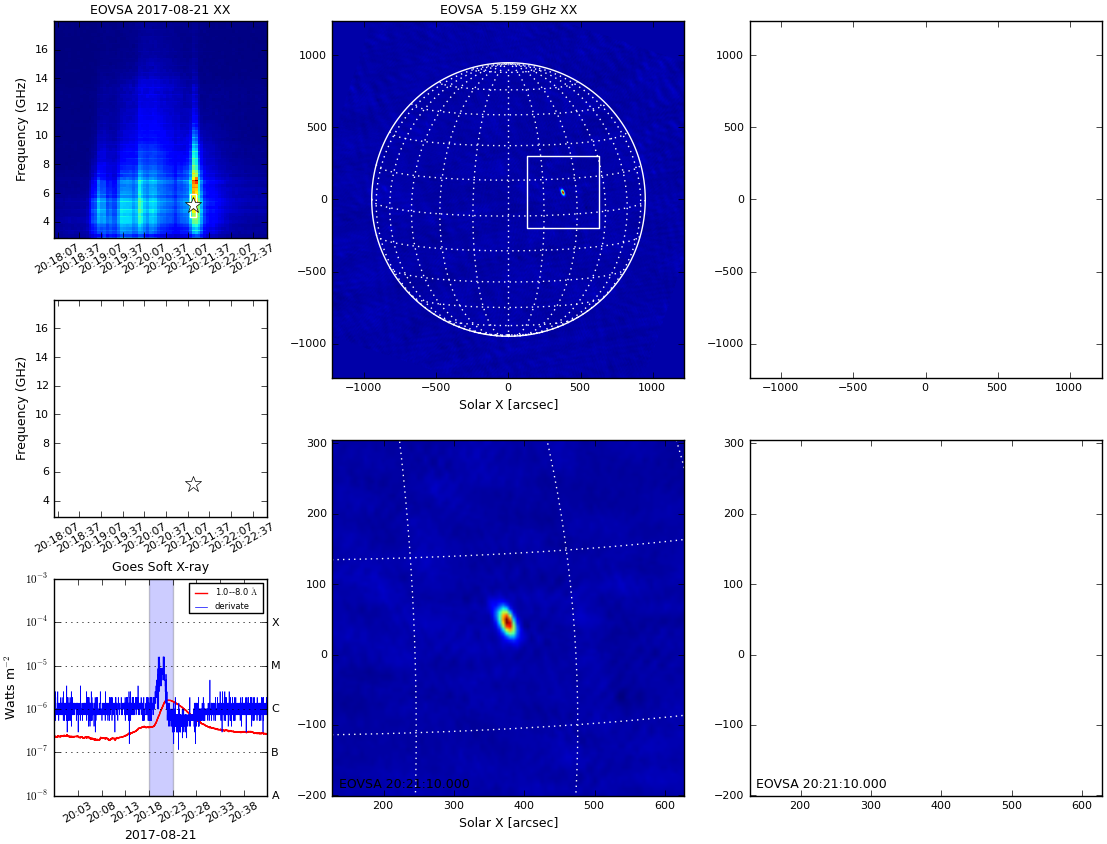
[[file:fig-qlookplot0.png|thumb|right|300px| | [[file:fig-qlookplot0.png|thumb|right|300px|EOVSA full-Sun single-band quicklook image]] | ||
<pre style="background-color: #FCEBD9"> | <pre style="background-color: #FCEBD9"> | ||
## in SunCASA | ## in SunCASA | ||
from suncasa.utils import qlookplot as ql | from suncasa.utils import qlookplot as ql | ||
msfile = 'IDB20170821201800-202300.4s.slfcaled.ms' | msfile = 'IDB20170821201800-202300.4s.slfcaled.ms' | ||
## | ## (Optional) Supply the npz file of the dynamic spectrum from previous step. | ||
## If not provided, the program will generate a new one from the visibility. | |||
specfile = msfile + '.dspec.npz' | specfile = msfile + '.dspec.npz' | ||
## set the time interval | ## set the time interval | ||
timerange = '20:21:10~20:21: | timerange = '20:21:10~20:21:18' | ||
## select | ## select frequency range from 5 GHz to 6 GHz | ||
spw = '5' | spw = '5~6GHz' | ||
## select stokes XX | ## select stokes XX | ||
stokes = 'XX' | stokes = 'XX' | ||
## turn off AIA image plotting, default is True | ## turn off AIA image plotting, default is True | ||
plotaia = False | plotaia = False | ||
ql.qlookplot(vis=msfile, specfile=specfile, timerange=timerange, spw=spw, \ | ql.qlookplot(vis=msfile, specfile=specfile, timerange=timerange, spw=spw, \ | ||
stokes=stokes | stokes=stokes, plotaia=plotaia) | ||
</pre> | </pre> | ||
With [https://github.com/ | |||
[[file:fig-qlookplot1.png|thumb|right|300px| | The empty panels are there as we only selected one polarization for imaging/spectroscopy. Feel free to explore by adjusting the above parameters, e.g. use a different time range ("timerange" parameter) and/or frequency range ("spw" parameter). | ||
With [https://github.com/suncasa/suncasa/blob/master/utils/qlookplot.py ''qlookplot''], it is easy to engage solar data from SDO/AIA in the summary plot. Setting ''plotaia=True'' in the [https://github.com/suncasa/suncasa/blob/master/utils/qlookplot.py ''qlookplot''] command will download SDO/AIA data at the given time to current directory and add it to the summary plot. | |||
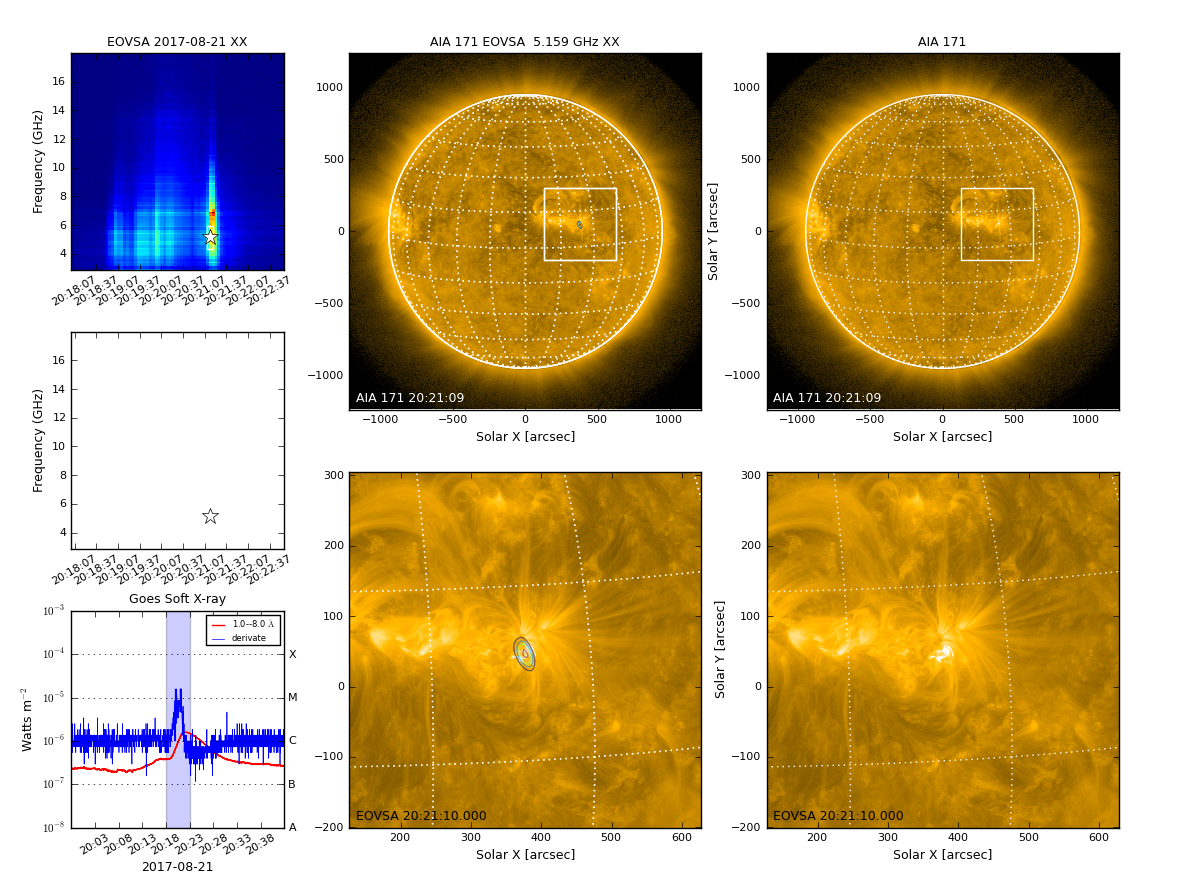
[[file:fig-qlookplot1.png|thumb|right|300px|EOVSA full-Sun single-band quicklook image with SDO/AIA as background]] | |||
<pre style="background-color: #FCEBD9"> | <pre style="background-color: #FCEBD9"> | ||
## in SunCASA | ## in SunCASA | ||
ql.qlookplot(vis=msfile, specfile=specfile, timerange=timerange, spw=spw, \ | ql.qlookplot(vis=msfile, specfile=specfile, timerange=timerange, spw=spw, \ | ||
stokes=stokes | stokes=stokes, plotaia=True) | ||
</pre> | </pre> | ||
The | The resulting radio image is a 4-D datacube (in solar X-pos, Y-pos, frequency, and polarization), which is, by default, saved as a fits file ''msfile + '.outim.image.fits''' under your working directory. The name of the output fits file can be specified using the "outfits" parameter. In this example, we combine all selected frequencies (specified in keyword "spw") into one image (i.e., multi-frequency synthesis). Therefore the third axis only has one plane. The fourth axis contains polarization. In this example, however, only the first one is populated ("XX"). Here is the relevant information in the header of the resulting FITS file. | ||
<pre> | <pre> | ||
NAXIS = 4 | NAXIS = 4 | ||
NAXIS1 = 512/ Nx | NAXIS1 = 512/ Nx | ||
| Line 161: | Line 169: | ||
NAXIS4 = 2/ number of polarization | NAXIS4 = 2/ number of polarization | ||
</pre> | </pre> | ||
By default, [https://github.com/ | By default, [https://github.com/suncasa/suncasa/blob/master/utils/qlookplot.py ''qlookplot''] produces a full sun radio image (512x512 with a pixel size of 5"). If you know where the radio source is (e.g., from the previous full-Sun imaging), you can make a partial solar image around the source by specifying the image center ("xycen"), pixel scale ("cell"), and image field of view ("fov"). Here we show an example that images a 8-s interval around 20:21:14 UT using multi-frequency synthesis in 12-14 GHz and a smaller restoring beam. The microwave source is show to bifurcate into two components, which correspond pretty well with the double flare ribbons in SDO/AIA. | ||
[[file:fig- | [[file:fig-qlookplot3.png|thumb|right|300px|EOVSA multi-frequency synthesis quicklook image]] | ||
<pre style="background-color: #FCEBD9"> | <pre style="background-color: #FCEBD9"> | ||
## in SunCASA | ## in SunCASA | ||
xycen = [375, 45] ## image center for clean in solar X-Y in arcsec | xycen = [375, 45] ## image center for clean in solar X-Y in arcsec | ||
cell=[ | cell=['2.0arcsec'] ## pixel scale | ||
imsize=[ | imsize=[128] ## number of pixels in X and Y. If only one value is provided, NX = NY | ||
fov = [100,100] ## field of view of the zoomed-in panels in unit of arcsec | fov = [100,100] ## field of view of the zoomed-in panels in unit of arcsec | ||
ql.qlookplot(vis=msfile, specfile=specfile, timerange= | ql.qlookplot(vis=msfile, specfile=specfile, timerange='20:21:10~20:21:18', \ | ||
restoringbeam= | spw='12GHz~14GHz', stokes='XX', restoringbeam=['6arcsec'],\ | ||
imsize=imsize,cell=cell,xycen=xycen,fov=fov,calpha=1.0) | |||
</pre> | </pre> | ||
Next, we will make | Next, we will make images for every single spectral window in this data set (from spw 1 to 30, spw 0 is in the visibility data, but is not calibrated for this event). | ||
[[file:fig-qlookplot_mbds.png|thumb|right|300px| | [[file:fig-qlookplot_mbds.png|thumb|right|300px|EOVSA multi-band quicklook image]] | ||
<pre style="background-color: #FCEBD9"> | <pre style="background-color: #FCEBD9"> | ||
## in SunCASA | ## in SunCASA | ||
xycen = [375, 45] ## image center for clean in solar X-Y in arcsec | xycen = [375, 45] ## image center for clean in solar X-Y in arcsec | ||
cell=[ | cell=['2.0arcsec'] ## pixel size | ||
imsize=[ | imsize=[128] ## x and y image size in pixels. | ||
fov = [100,100] ## field of view of the zoomed-in panels in unit of arcsec | fov = [100,100] ## field of view of the zoomed-in panels in unit of arcsec | ||
spw = ['{}'.format( | spw = ['{}'.format(s) for s in range(1,31)] | ||
clevels = [0.5, 1.0] ## | clevels = [0.5, 1.0] ## contour levels to fill in between. | ||
calpha=0.35 ## the alpha | calpha=0.35 ## now tune down the alpha | ||
restoringbeam=['6arcsec'] | |||
ql.qlookplot(vis=msfile, specfile=specfile, timerange=timerange, spw=spw, stokes=stokes, \ | ql.qlookplot(vis=msfile, specfile=specfile, timerange=timerange, spw=spw, stokes=stokes, \ | ||
restoringbeam=restoringbeam,imsize=imsize,cell=cell, \ | restoringbeam=restoringbeam,imsize=imsize,cell=cell, \ | ||
xycen=xycen,fov=fov,clevels=clevels,calpha=calpha) | xycen=xycen,fov=fov,clevels=clevels,calpha=calpha) | ||
</pre> | </pre> | ||
The output | The output FITS file as a 4-d cube is saved to ''msfile + '.outim.image.fits''' under your working directory. The 30 spectral windows used for spectral imaging are combined in the output FITS file. So NAXIS3 (frequency axis) is 30. | ||
<pre> | <pre> | ||
NAXIS = 4 / number of array dimensions | NAXIS = 4 / number of array dimensions | ||
NAXIS1 = 128 | NAXIS1 = 128 | ||
| Line 197: | Line 205: | ||
NAXIS3 = 30 | NAXIS3 = 30 | ||
NAXIS4 = 2 | NAXIS4 = 2 | ||
</pre> | |||
===Quick-Look Imaging series=== | |||
<pre style="background-color: #FCEBD9;"> | |||
# In SunCASA | |||
msfile = 'IDB20170821201800-202300.4s.slfcaled.ms' | |||
specfile = msfile + '.dspec.npz' | |||
## set the time interval | |||
timerange = '2017/08/21/20:21:10~2017/08/21/20:21:18' | |||
## Bin width for time averaging | |||
twidth = 1 | |||
## frequency range | |||
spw = ['0~3'] | |||
## image center for clean in solar X-Y in arcsec | |||
xycen = [375, 45] | |||
## number of pixels in X and Y. If only one value is provided, NX = NY | |||
imsize = [128] | |||
## field of view of the zoomed-in panels in unit of arcsec | |||
fov = [100., 100.] | |||
## pixel scale | |||
cell = ['2.0arcsec'] | |||
## select stokes XX | |||
stokes = 'XX' | |||
## for EOVSA data, set usemsphacenter to False | |||
usemsphacenter = False | |||
## set True if make a movie | |||
mkmovie = True | |||
## set True if generate compressed fits | |||
docompress = True | |||
#### ---- Control knobs for AIA plotting ---- #### | |||
## set True if plot AIA images as the background | |||
plotaia = True | |||
## provide the path to the directory where the AIA fits files are located. Otherwise, set it to be '' | |||
aiadir = 'Directory-where-AIA-fits-files-are-located' | |||
## AIA passband. The options are [171,131,304,335,211,193,94,1600,1700] | |||
aiawave = 171 | |||
## numbers of CPU threads for computing | |||
ncpu = 2 | |||
ql.qlookplot(vis=msfile, specfile=specfile, timerange=timerange, spw=spw, | |||
xycen=xycen, imsize=imsize, fov=fov, cell=cell, usemsphacenter=usemsphacenter, | |||
plotaia=plotaia, aiadir=aiadir, aiawave=aiawave, | |||
mkmovie=mkmovie, twidth=twidth, ncpu=ncpu, docompress=docompress, stokes=stokes) | |||
</pre> | </pre> | ||
===Batch-Mode Imaging=== | ===Batch-Mode Imaging=== | ||
This section is for interested users who wish to generate FITS files with full control on all parameters being used for synthesis imaging. We provide one example SunCASA script for generating 30-band spectral imaging maps, and another for iterating over time to produce a time series of these maps. | This section is for interested users who wish to generate FITS files with full control on all parameters being used for synthesis imaging. We provide one example SunCASA script for generating 30-band spectral imaging maps, and another for iterating over time to produce a time series of these maps. | ||
====Producing a 30-band map | ====Producing a 30-band map cube for a given time==== | ||
An example script can be found at [https://github.com/binchensun/eovsa-tutorial/blob/master/rhessi18/imaging_example.py this Github link]. If you are on the AWS server Virgo, it is under /common/data/eovsa_tutorial/imaging_example.py. First, download or copy the script to your own working directory and cd to your directory. | An example script can be found at [https://github.com/binchensun/eovsa-tutorial/blob/master/rhessi18/imaging_example.py this Github link]. If you are on the AWS server Virgo, it is under /common/data/eovsa_tutorial/imaging_example.py. First, download or copy the script to your own working directory and cd to your directory. | ||
<pre> | <pre style="background-color:#FCEBD9;"> | ||
cd your_working_directory | # in SunCASA | ||
cp /common/data/eovsa_tutorial/imaging_example.py ./ | CASA <##>: cd your_working_directory | ||
CASA <##>: !cp /common/data/eovsa_tutorial/imaging_example.py ./ | |||
</pre> | </pre> | ||
| Line 211: | Line 266: | ||
Second (optional), change inputs in the following block in your copy of the "imaging_example.py" script and save the changes. This block has definitions for time range, image center and FOV, antennas used, cell size, number of pixels, etc. | Second (optional), change inputs in the following block in your copy of the "imaging_example.py" script and save the changes. This block has definitions for time range, image center and FOV, antennas used, cell size, number of pixels, etc. | ||
<pre | <pre> | ||
################### USER INPUT GOES IN THIS BLOK | ################### USER INPUT GOES IN THIS BLOK ################ | ||
vis = 'IDB20170821201800-202300.4s.slfcaled.ms' # input visibility data | vis = 'IDB20170821201800-202300.4s.slfcaled.ms' # input visibility data | ||
trange = '2017/08/21/20:21:00~2017/08/21/20:21:30' # | trange = '2017/08/21/20:21:00~2017/08/21/20:21:30' # time range for imaging | ||
xycen = [380., 50.] # | xycen = [380., 50.] # center of the output map (in solar X and Y, Unit: arcsec) | ||
xran = [ | xran = [340., 420.] # plot range in solar X. Unit: arcsec | ||
yran = [ | yran = [10., 90.] # plot range in solar Y. Unit: arcsec | ||
antennas = ' | antennas = '' # Use all 13 EOVSA 2-m antennas by default | ||
npix = 256 # number of pixels in the image | npix = 256 # number of pixels in the image | ||
cell = '1arcsec' # pixel scale in arcsec | cell = '1arcsec' # pixel scale in arcsec | ||
pol = 'XX' # polarization to image, use XX for now | pol = 'XX' # polarization to image, use XX for now | ||
pbcor = True # correct for primary beam response? | pbcor = True # correct for primary beam response? | ||
outdir = './images' # Specify where | outdir = './images' # Specify where to save the output fits files | ||
outimgpre = 'EO' # Something to add to the output image name | outimgpre = 'EO' # Something to add to the output image name | ||
</pre> | </pre> | ||
Then, run the script in SunCASA | Then, run the script in SunCASA via | ||
<pre style="background-color:#FCEBD9;"> | <pre style="background-color:#FCEBD9;"> | ||
execfile('imaging_example.py') | CASA <##>: execfile('imaging_example.py') | ||
</pre> | </pre> | ||
The output | The output is a combined 30-band FITS file saved under "outdir". The naming conversion is outimgpre + YYYYMMDDTHHMMSS_ALLBD.fits. In this example, it is "./images/EO20170821T202115.000_ALLBD.fits". Here is the detailed information of the axes of the output FITS file. | ||
<pre> | |||
NAXIS = 4 / number of array dimensions | |||
EO20170821T202115. | NAXIS1 = 128 | ||
NAXIS2 = 128 | |||
NAXIS3 = 30 | |||
NAXIS4 = 2 | |||
</pre> | </pre> | ||
From this point, you can use your favorite language | From this point, you can use your favorite language to read the fits files and plot them. The last block of the example script uses SunPy.map to generate the plot shown on the right. For SSWIDL users, please refer to [[#Plotting Images in SSWIDL|Section 3.3.3.3]]. | ||
====Producing a time series of 30-band maps (a 4-d cube)==== | |||
An example script can be found at [https://github.com/binchensun/eovsa-tutorial/blob/master/rhessi18/imaging_timeseries_example.py this Github link]. If you are on the AWS server Virgo, it is under /common/data/eovsa_tutorial/imaging_timeseries_example.py. The script enables parallel-processing (based on a customized task "ptclean3" -- do not ask me why we have "3" in the task name). To invoke more CPU processes for parallel-processing, change "nthreads" in the preambles from 1 to the number of threads you wish to use. | |||
<span style="color:#ff0000">Caution: This script is very computationally intensive. '''Please do not try to run this script on the AWS server during the RHESSI 18 live tutorial''', as it has limited resources that everyone must share. </span> | |||
<pre> | <pre style="background-color:#FCEBD9;"> | ||
cd your_working_directory | # in SunCASA | ||
cp | CASA <##>: cd your_working_directory | ||
CASA <##>: !cp /common/data/eovsa_tutorial/imaging_timeseries_example.py ./ | |||
</pre> | </pre> | ||
Similar as the previous script, change inputs in the following | Similar as the previous script, change inputs in the following block of your copy of the "imaging_timeseries_example.py" script and save the changes. | ||
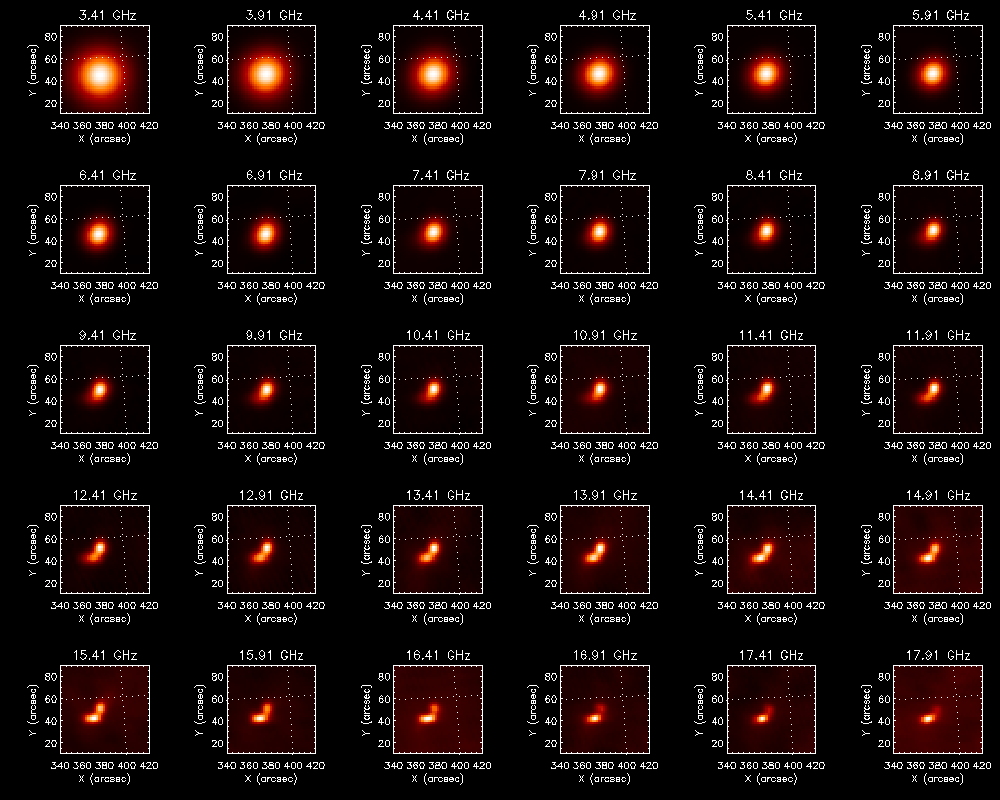
[[file:fig-specmovie.png|thumb|right|300px|Example multi-frequency images at one time frame in the output movie]] | [[file:fig-specmovie.png|thumb|right|300px|Example multi-frequency images at one time frame in the [https://web.njit.edu/~sjyu/download/eovsa-tutorial/movie.html output javascript movie]]] | ||
<pre | <pre> | ||
################### USER INPUT GOES IN THIS BLOK | ################### USER INPUT GOES IN THIS BLOK #################### | ||
vis = 'IDB20170821201800-202300.4s.slfcaled.ms' # input visibility data | vis = 'IDB20170821201800-202300.4s.slfcaled.ms' # input visibility data | ||
specfile = vis + '.dspec.npz' ## input dynamic spectrum | specfile = vis + '.dspec.npz' ## input dynamic spectrum | ||
nthreads = 1 # Number of processing threads to use | nthreads = 1 # Number of processing threads to use | ||
overwrite = True # whether to overwrite the existed fits files. | overwrite = True # whether to overwrite the existed fits files. | ||
trange = '' # | trange = '' # time range for imaging, default to all times in the data | ||
twidth = | twidth = 1 # make one image out of every 2 time integrations | ||
xycen = [380., 50.] # | xycen = [380., 50.] # center of the output map in solar X and Y. Unit: arcsec | ||
xran = [ | xran = [340., 420.] # plot range in solar X. Unit: arcsec | ||
yran = [ | yran = [10., 90.] # plot range in solar Y. Unit: arcsec | ||
antennas = ' | antennas = '' # default is to use all 13 EOVSA 2-m antennas. | ||
npix = 128 # number of pixels in the image | npix = 128 # number of pixels in the image | ||
cell = '2arcsec' # pixel scale in arcsec | cell = '2arcsec' # pixel scale in arcsec | ||
pol = 'XX' # polarization to image, use XX for now | pol = 'XX' # polarization to image, use XX for now | ||
pbcor = True # correct for primary beam response? | pbcor = True # correct for primary beam response? | ||
grid_spacing = 5. * u.deg # spacing | grid_spacing = 5. * u.deg # grid spacing in degrees | ||
outdir = './image_series/' # Specify where | outdir = './image_series/' # Specify where to save the output fits files | ||
imresfile = 'imres.npz' # File to write the | imresfile = 'imres.npz' # File to write summary of the imaging results | ||
outimgpre = 'EO' # Something to add to the image name | outimgpre = 'EO' # Something to add to the image name | ||
</pre> | </pre> | ||
Then, run the script in SunCASA by | Then, run the script in SunCASA by | ||
<pre style="background-color:#FCEBD9;"> | <pre style="background-color:#FCEBD9;"> | ||
execfile('imaging_timeseries_example.py') | CASA <##>: execfile('imaging_timeseries_example.py') | ||
</pre> | |||
The output fits images and a summary file imresfile are saved under "outdir" (in this example "./image_timeseries"). The naming convention of output fits images is the same as [[#Producing a 30-band map at a given time|the previous section]]. The summary yields the time, frequency, and path to every image. | |||
<pre style="background-color:#FCEBD9;"> | |||
CASA <##>: imres = np.load(outdir + imresfile)['imres'].item() | |||
CASA <##>: imres.keys() | |||
['FreqGHz', 'ImageName', 'Succeeded', 'BeginTime', 'EndTime'] | |||
CASA <##>: ls image_series/*.fits | head -5 | |||
image_series/EO20170821T201800.500_ALLBD.fits | |||
image_series/EO20170821T201804.500_ALLBD.fits | |||
image_series/EO20170821T201808.500_ALLBD.fits | |||
image_series/EO20170821T201812.500_ALLBD.fits | |||
image_series/EO20170821T201816.500_ALLBD.fits | |||
</pre> | |||
The last block of the example script uses SunPy.map to generate a series of plots and wraps them as a [https://web.njit.edu/~sjyu/download/eovsa-tutorial/movie.html javascript movie]. | |||
====Plotting Images in SSWIDL==== | |||
All the output files are in standard FITS format in the Helioprojective Cartesian coordinate system (that most spacecraft solar image data adopt; FITS header CTYPE is HPLN-TAN and HPLT-TAN). They are fully compatible with [https://hesperia.gsfc.nasa.gov/rhessidatacenter/complementary_data/maps/ the SSWIDL map suite] which deals with FITS files. We have prepared SSWIDL routines to convert the single time or time-series FITS files to an array of SSWIDL map structure. The scripts are available in the [https://github.com/binchensun/eovsa-tutorial/tree/master/rhessi18 Github repository of our tutorial]. For those working on Virgo, local copies are placed under /common/data/eovsa_tutorial/. Two scripts are relevant to this tutorial: | |||
* [https://github.com/binchensun/eovsa-tutorial/blob/master/rhessi18/casa_readfits.pro casa_readfits.pro]: read an array of multi-frequency FITS files into FITS header (index) and data. | |||
* [https://github.com/binchensun/eovsa-tutorial/blob/master/rhessi18/casa_fits2map.pro casa_fits2map.pro]: convert an array of multi-frequency FITS files into an array of SSWIDL map structure (adapted from P. T. Gallagher's hsi_fits2map.pro) | |||
First, start SSWIDL from command line: | |||
<pre style="background-color:#FCEBD9;"> | |||
sswidl | |||
</pre> | |||
Find and convert the fits files: | |||
<pre style="background-color:#FCEBD9;"> | |||
; cd to your path that contains casa_readfits.pro and casa_fits2map.pro. If on Virgo, just add the path | |||
IDL> add_path, '/common/data/eovsa_tutorial/' | |||
IDL> fitsdir = '/common/data/eovsa_tutorial/image_series/' | |||
IDL> fitsfiles = file_search(fitsdir+'*_ALLBD.fits') | |||
; The resulting fitfiles contains all multi-frequency FITS files under "fitsdir" | |||
; The following command converts the fits files to an array of map structures. | |||
; "casa_fits2map.pro" also work well on a single FITS file. | |||
; (Optional) keyword "calcrms" is to calculate rms and dynamic range (SNR) of the maps in a user-defined "empty" region | |||
; of the map specified by "rmsxran" and "rmsyran" | |||
;;; Here we load 10 time frames as a demonstration | |||
IDL> casa_fits2map, fitsfiles[45:54], eomaps [, /calcrms, rmsxran=[260., 320.], rmsyran=[-70., 0.]] | |||
</pre> | </pre> | ||
The | The resulting maps have a shape of [# of frequencies, # of polarizations, # of times] | ||
<pre> | <pre style="background-color:#FCEBD9;"> | ||
IDL> help,eomaps | |||
EOMAPS STRUCT = -> <Anonymous> Array[30, 1, 10] | |||
</pre> | |||
They have compatible keywords recognized by, e.g., plot_map.pro. Example of the output map keywords: | |||
<pre style="background-color:#FCEBD9;"> | |||
IDL> help,eomaps,/str | |||
** Structure <36009f8>, 24 tags, length=131336, data length=131332, refs=1: | |||
(' | DATA DOUBLE Array[128, 128] | ||
XC DOUBLE 378.99175 | |||
YC DOUBLE 49.001472 | |||
DX DOUBLE 2.0000000 | |||
DY DOUBLE 2.0000000 | |||
TIME STRING '21-Aug-2017 20:20:59.500' | |||
ID STRING 'EOVSA XX 3.413GHz' | |||
DUR DOUBLE 3.9999997 | |||
XUNITS STRING 'arcsec' | |||
YUNITS STRING 'arcsec' | |||
ROLL_ANGLE DOUBLE 0.0000000 | |||
ROLL_CENTER DOUBLE Array[2] | |||
FREQ DOUBLE 3.4125694 | |||
FREQUNIT STRING 'GHz' | |||
STOKES STRING 'XX' | |||
DATAUNIT STRING 'K' | |||
DATATYPE STRING 'Brightness Temperature' | |||
BMAJ DOUBLE 0.0097377778 | |||
BMIN DOUBLE 0.0097377778 | |||
BPA DOUBLE 0.0000000 | |||
RSUN DOUBLE 948.03989 | |||
L0 FLOAT 0.00000 | |||
B0 DOUBLE 6.9299225 | |||
COMMENT STRING 'Converted by CASA_FITS2MAP.PRO' | |||
</pre> | |||
[[file:fig-specimg_idl.png|thumb|right|300px|Example multi-frequency EOVSA images at one time plotted in SSWIDL]] | |||
An example for plotting all images at a selected time: | |||
<pre style="background-color:#FCEBD9;"> | |||
; choose a time pixel | |||
IDL> plttime = anytim('2017-08-21T20:21:15') | |||
IDL> dt = min(abs(anytim(eomaps[0,0,*].time)-plttime),tidx) | |||
; use maps at this time, first polarization, and all bands | |||
IDL> eomap = reform(eomaps[*,0,tidx]) | |||
IDL> window,0,xs=1000,ys=800 | |||
IDL> !p.multi=[0,6,5] | |||
IDL> loadct, 3 | |||
IDL> for i=0, 29 do plot_map,eomap[i],grid=5.,$ | |||
tit=string(eomap[i].freq,format='(f5.2)')+' GHz', $ | |||
chars=2.0,xran=[340.,420.],yran=[10.,90.] | |||
</pre> | </pre> | ||
==Spectral Fitting with GSFIT== | ==Spectral Fitting with GSFIT== | ||
In this section we discuss using GSFIT (now a package of SSWIDL) to perform spectral fit based on the resulting FITS images produced from the previous steps. We will use the FITS images produced in [[#Producing a time series of 30-band maps (a 4-d cube)|Section 3.3.3.2]] as the input. On Virgo, we prepared an IDL save file located at /common/data/eovsa_tutorial/21August2017.sav. In addition, this folder contains three more demo files that may be used to demonstrate the GSFIT functionality. | |||
For those intending to use their own machines for this part of the tutorial, these IDL fits files may be downloaded from [https://www.dropbox.com/sh/tjez92wfph1jp7x/AAAJBO6AJ0GC5H3GCZypc2BYa?dl=0 this location] | |||
===GSFIT GUI Application=== | ===GSFIT GUI Application=== | ||
The GSFIT GUI application may be launched as follows | The GSFIT GUI application may be launched as follows | ||
Latest revision as of 18:02, 20 May 2022
Browsing and Obtaining EOVSA data
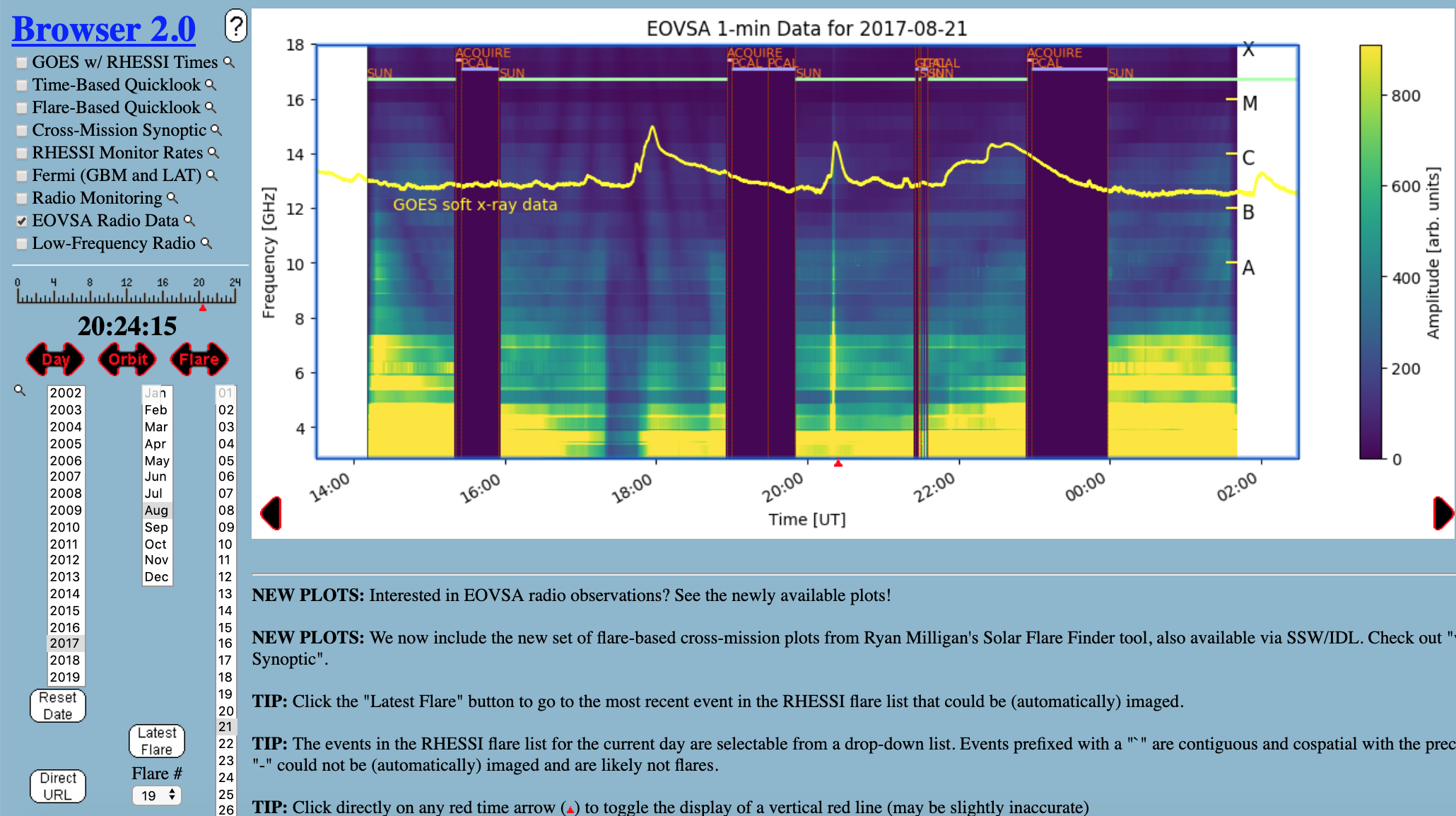
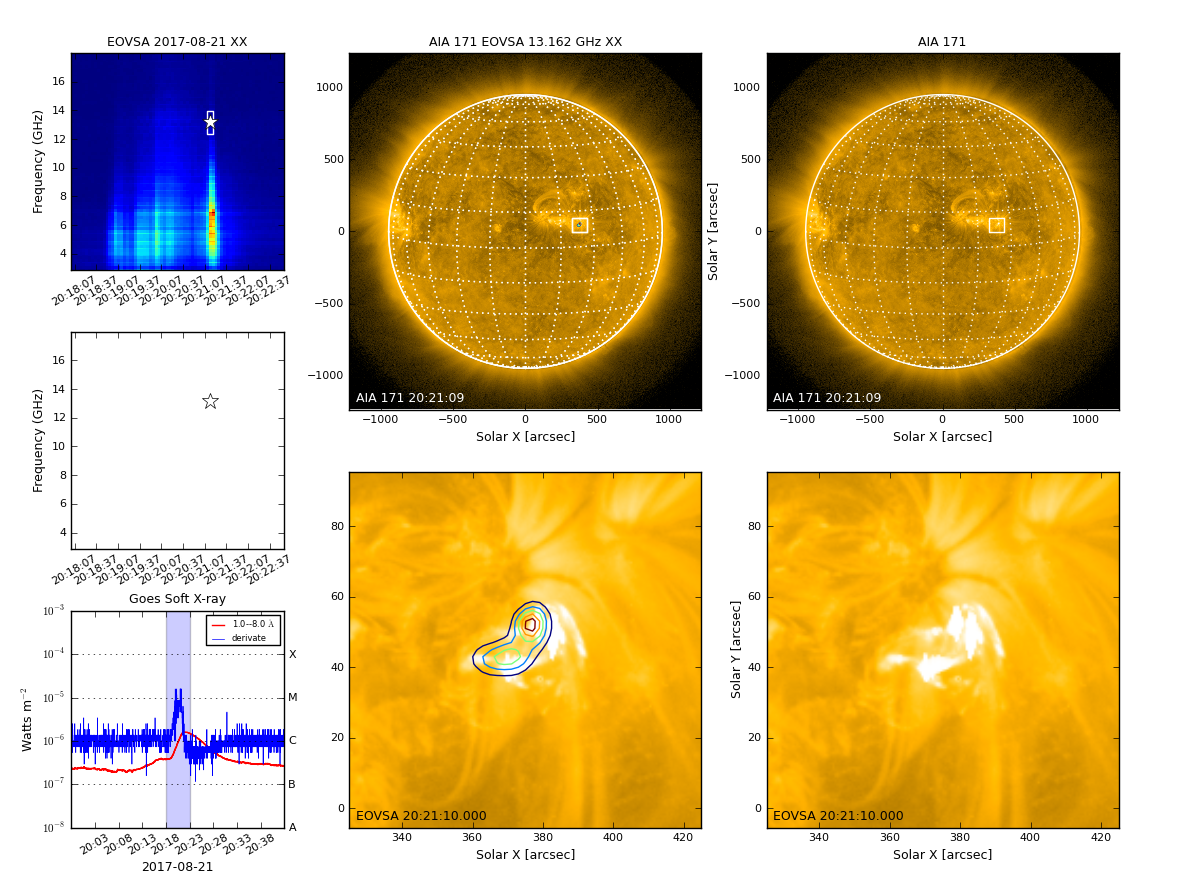
Currently the most convenient way for browsing EOVSA data is through the RHESSI Browser. First, check the "EOVSA Radio Data" box on the data selection area (top-left corner). Then select year/month/date to view the overall EOVSA dynamic spectrum. Note if a time is selected at early UTC hours (e.g., 0-3 UT), it will show the EOVSA dynamic spectrum from the previous day. Also note that EOVSA data were not commissioned for spectroscopic imaging prior to April 2017. An example of the overview EOVSA dynamic spectrum for 2017 Aug 21 is shown in the figure on the right.
The overview EOVSA dynamic spectra are from the median of the uncalibrated cross-power visibilities at a few short baselines, which are not (but a good proxy of) the total-power dynamic spectra. The effects of spatial information blended in the cross-power visibilities can be clearly seen as the "U"-shaped features throughout the day, which are due to the movement of the Sun across the sky that effectively changes the length and orientation of the baselines. Flare times can be easily seen in the EOVSA dynamic spectra, which usually appear as vertical bright features across many frequency bands. More information can be found on this page.
We are currently working on a pipeline to create quicklook full-disk images for implementation at the RHESSI Browser in a way similar as the RHESSI quicklook images. Please stay tuned.
Once you identify the flare time, you can find the full-resolution (1-s cadence) uncalibrated visibility files (in Miriad format) at this link. Each data file is usually 10 minutes in duration. Name convention is YYYYMMDD/IDBYYYYMMDDHHMMSS, where the time in the file name indicates the start time of the visibility data.
Calibrating EOVSA data
- Calibration: Calibrating EOVSA data is an involved process. If you are interested the Practical Calibration Tutorial describes the procedure. We are currently working on a semi-automatic pipeline for calibrating the visibility data. At this moment, however, please contact the EOVSA team if you wish to have calibrated visibility data for a specific event. In the near future, we envision the pipeline-calibrated visibility data to be available for download on a regular basis.
- Self-calibration: For improving the quality of the imaging, self-calibration is usually needed. This is still a work-in-progress, but we have had some successful practice in self-calibrating some flare events. Please refer to Self-Calibrating Flare Data Tohban_EOVSA_Imaging_Tutorial_A-Z for examples and detailed discussions on our current practice for self-calibrating EOVSA flare data.
Analyzing EOVSA data
Software
We have developed two packages for EOVSA data processing and analysis:
- SunCASA A wrapper around CASA (the Common Astronomy Software Applications package) for synthesis imaging and visualizing solar spectral imaging data. CASA is one of the leading software tool for "supporting the data post-processing needs of the next generation of radio astronomical telescopes such as ALMA and VLA", an international effort led by the National Radio Astronomy Observatory. The current version of CASA uses Python (2.7) interface. More information about CASA can be found on NRAO's CASA website . Note, CASA is available ONLY on UNIX-BASED PLATFORMS (and therefore, so is SunCASA).
- GSFIT A IDL-widget(GUI)-based spectral fitting package called gsfit, which provides a user-friendly display of EOVSA image cubes and an interface to fast fitting codes (via platform-dependent shared-object libraries).
For this tutorial, there are two approaches in accessing our software packages:
- 1. Through our Amazon Cloud server. See Section 3.1.1 for detailed instructions.
- 2. Install on your own machine. See Section 3.1.2 (SunCASA) and Section 3.1.3 (GSFIT) for instructions.
Using Software on Amazon Cloud Server
We use an Amazon AWS Lightsail cloud server for lightweight data processing and testing purposes. The server has 2 CPUs, 8 GB RAM, and 160 GB SSD storage. It runs CentOS 7 (1901-01) Linux.
NOTE: the guest account is ONLY INTENDED FOR THE EOVSA TUTORIAL, WHICH WILL BE DISABLED SHORTLY AFTER THE RHESSI WORKSHOP (you will be notified prior to that). If you wish to continue using the server for testing purposes after the tutorial, please contact Bin Chen to set up a personal account.
DISCLAIMER: We are NOT RESPONSIBLE for any loss, damage, or leak of your personal data on the server.
- Obtain SSH Key to the guest account from Bin Chen
- Put it under a secure location on your own machine.
- Follow remaining directions depending on your client machine.
Connecting via command line
- Edit the permission of the SSH key and the directory you choose to place it. Here I use ~/.ssh as an example (create it first by mkdir ~/.ssh if it does not exist.)
chmod 700 ~/.ssh chmod 400 ~/.ssh/guest-virgo.pem
- Log on to AWS server (password-less)
ssh -X -i ~/.ssh/guest-virgo.pem guest@virgo.arcs.az.njit.edu
Now you are connected to virgo.
Connecting via Windows (MobaXterm)
Recommend to use "Documents\MobaXterm\home\.ssh", which should exist if you have already installed the free MobaXterm[1].
- Create new session, click SSH, enter virgo.arcs.az.njit.edu for Remote host, guest for username.
- On advanced SSH settings tab, click Use private key, navigate to and select file guest_virgo.pem
- Close setup window and click the new session's icon, which will log you in.
Startup SunCASA and sswIDL
Once you are connected to virgo, you will have access to SunCASA and GSFIT (included in the sswIDL installation). To not interfere with others (who share the same "guest" account), please create your own directory and work under it. For easier identification, please kindly use the initial of your first name and your full last name as the name of your directory (here "bchen" is used as an example).
[guest@ip-172-26-5-203 ~]$ mkdir bchen [guest@ip-172-26-5-203 ~]$ cd bchen
To enter SunCASA
[guest@ip-172-26-5-203 ~/bchen]$ suncasa
To enter sswIDL
[guest@ip-172-26-5-203 ~/bchen]$ sswidl
SunCASA Installation
Please follow this link for details regarding installation of SunCASA on your own machine (only available on Unix-bases OS). This will take you to another page.
GSFIT Installation
Please follow this link for details regarding installation of GSFIT on your own machine (which supports multiple platforms). This will take you to another page.
Example Data and Scripts
In this tutorial, we will demonstrate steps for spectral imaging (self-)calibrated visibility data for a C-class flare on 2017 Aug 21 at ~20:20 UT (Section 3.3), followed by a tutorial on visualizing and analyzing the resulting spectral imaging cube (Section 3.4).
The visibility data can be accessed by
- Downloading from this Google Drive link.
- If you are on our AWS cloud server "virgo" (means for accessing the server are discussed in Section 3.1.1), it is located at /common/data/eovsa_tutorial/IDB20170821201800-202300.4s.slfcaled.ms.
For completeness, the pre-slfcalibration visibility data can be downloaded from this Google Drive link (not used by this tutorial).
To reduce the amount of data for this tutorial, the time resolution has been reduced by a factor of 4, to 4 s per sample. The actual time resolution available is 1 s. If you are on virgo, please copy it over to your own working directory that you created earlier under the guest account (e.g., ~/bchen/).
cd ''your_working_directory'' cp -r /common/data/eovsa_tutorial/IDB20170821201800-202300.4s.slfcaled.ms ./
Example scripts for doing the analysis covered in this tutorial are available as this Github repository. On Virgo, the scripts can be found under /common/data/eovsa_tutorial/.
Spectral Imaging with SunCASA
EOVSA data is handled in CASA tables system, known as a Measurement Set (MS). The actual visibility data are stored in a MAIN table that contains a number of rows, each of which is effectively a single timestamp for a single spectral window and a single baseline. Within SunCASA, in addition to everything already available in CASA, you have access to additional tools that allow you to explore and utilize the new radio dynamic spectroscopic imaging data from EOVSA. We also had success in using SunCASA for analyzing data from the Karl G. Jansky Very Large Array. The steps are very similar to those described here. To start SunCASA, use:
suncasa
Within SunCASA, you are under the IPython environment. Everything you know about (I)Python should be applicable here. The installation comes with frequently used packages including Matplotlib, Numpy, SciPy, AstroPy, SunPy. However, it is not very intuitive to add (compatible) Python packages within (Sun)CASA. If you choose to do so, you may want to check this page on how to install AstroPy (which is not originally shipped with CASA) within CASA. This method is generally applicable to adding other packages (but not thoroughly tested). If you need some specific packages for your analysis, and it does not require direct interaction with (Sun)CASA, we recommend you to use the standard Python environment. On Virgo, we have installed Anaconda 3, which can be accessed by, e.g., typing "ipython" in a terminal window.
Cross-Power Dynamic Spectrum
The first module we introduce is dspec. This module allows you to generate a cross-power dynamic spectrum from an MS file, and visualize it. You can select a subset of data by specifying a time range, spectral windows/channels, antenna baseline, or uvrange. The selection syntax follows the CASA convention. More information of CASA selection syntax may be found in the above links or the Measurement Selection Syntax.
from suncasa.utils import dspec as ds
import matplotlib.pyplot as plt
## define the visbility data file
msfile = 'IDB20170821201800-202300.4s.slfcaled.ms'
## define the output filename of the dynamic spectrum
specfile = msfile + '.dspec.npz'
## select relatively short baselines within a length (here I use 0.15~0.5km),
## and take a median cross all of them (with the domedian parameter)
## alternatively, you can use the "bl" parameter to select individual baseline(s)
uvrange = '0.15~0.5km'
## this step generates a dynamic spectrum and saves it to specfile
dspec=ds.get_dspec(vis=msfile, specfile=specfile, uvrange=uvrange, domedian=True)
## dspec is a Python dictionary that contains the resulting dynamic spectrum.
## A copy is saved in "specfile" as a numpy npz file.
## Other optional parameters are available for more selection criteria
## such as frequency range ("spw"), and time range ("timeran")
## Use "ds.get_dspec?" to see more options
## define the polarizations to show (here I use XX and YY)
pol='XXYY'
## The following command displays the resulting cross-power dynamic spectrum
ds.plt_dspec(specfile, pol=pol) # alternatively you can use the output from the previous step "dspec" as the input
Now you should have a popup window showing the dynamic spectrum. Hover your mouse over the dynamic spectrum, you can read the time and frequency information at the bottom of the window.
Quick-Look Imaging
Imaging EOVSA data involves image cleaning, as well as solar coordinate transformation and image registration. We bundled a number of these steps ino a module named qlookplot, allowing users to generate an observing summary plot showing the cross power dynamic spectrum, GOES light curves and EOVSA quick-look images. Now let us start with making a summary plot of EOVSA image at the spectral window 5 (5.4 GHz).
## in SunCASA
from suncasa.utils import qlookplot as ql
msfile = 'IDB20170821201800-202300.4s.slfcaled.ms'
## (Optional) Supply the npz file of the dynamic spectrum from previous step.
## If not provided, the program will generate a new one from the visibility.
specfile = msfile + '.dspec.npz'
## set the time interval
timerange = '20:21:10~20:21:18'
## select frequency range from 5 GHz to 6 GHz
spw = '5~6GHz'
## select stokes XX
stokes = 'XX'
## turn off AIA image plotting, default is True
plotaia = False
ql.qlookplot(vis=msfile, specfile=specfile, timerange=timerange, spw=spw, \
stokes=stokes, plotaia=plotaia)
The empty panels are there as we only selected one polarization for imaging/spectroscopy. Feel free to explore by adjusting the above parameters, e.g. use a different time range ("timerange" parameter) and/or frequency range ("spw" parameter).
With qlookplot, it is easy to engage solar data from SDO/AIA in the summary plot. Setting plotaia=True in the qlookplot command will download SDO/AIA data at the given time to current directory and add it to the summary plot.
## in SunCASA
ql.qlookplot(vis=msfile, specfile=specfile, timerange=timerange, spw=spw, \
stokes=stokes, plotaia=True)
The resulting radio image is a 4-D datacube (in solar X-pos, Y-pos, frequency, and polarization), which is, by default, saved as a fits file msfile + '.outim.image.fits' under your working directory. The name of the output fits file can be specified using the "outfits" parameter. In this example, we combine all selected frequencies (specified in keyword "spw") into one image (i.e., multi-frequency synthesis). Therefore the third axis only has one plane. The fourth axis contains polarization. In this example, however, only the first one is populated ("XX"). Here is the relevant information in the header of the resulting FITS file.
NAXIS = 4 NAXIS1 = 512/ Nx NAXIS2 = 512/ Ny NAXIS3 = 1/ number of frequency NAXIS4 = 2/ number of polarization
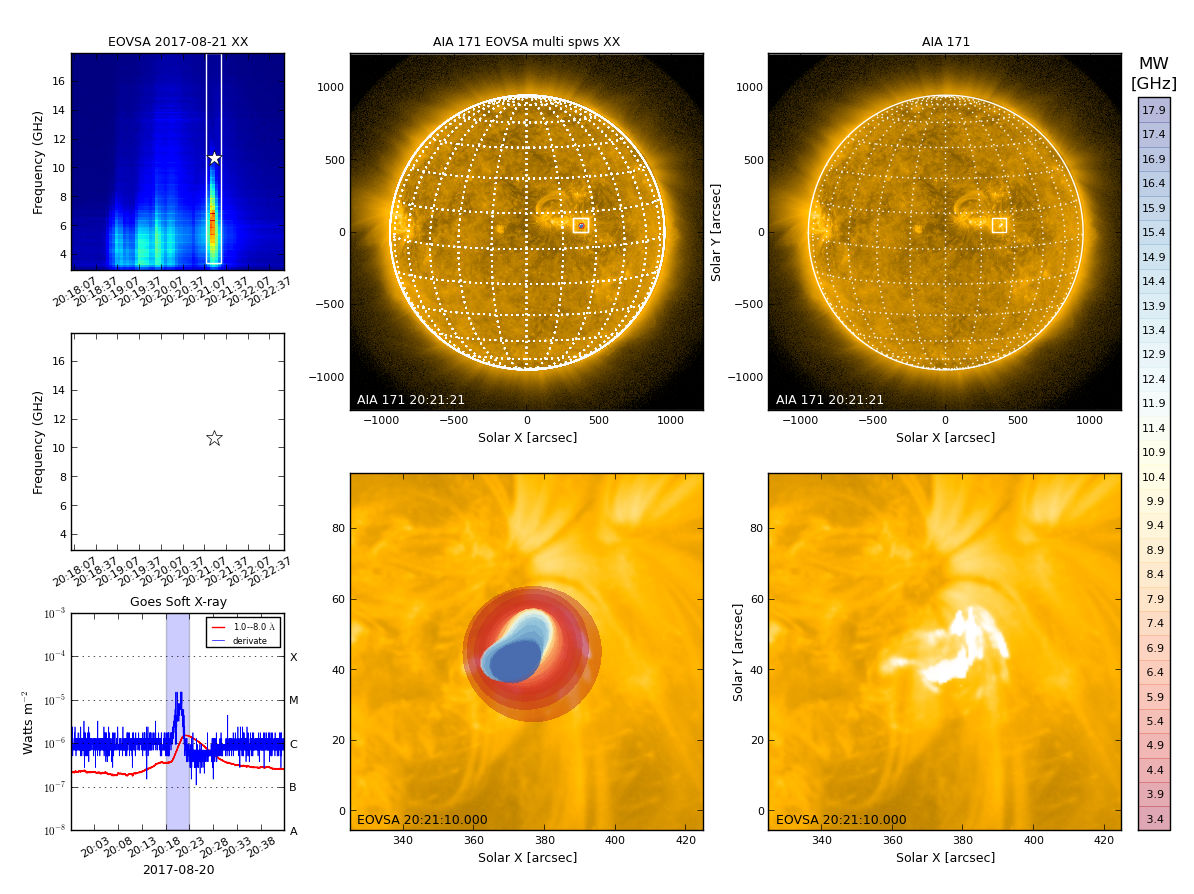
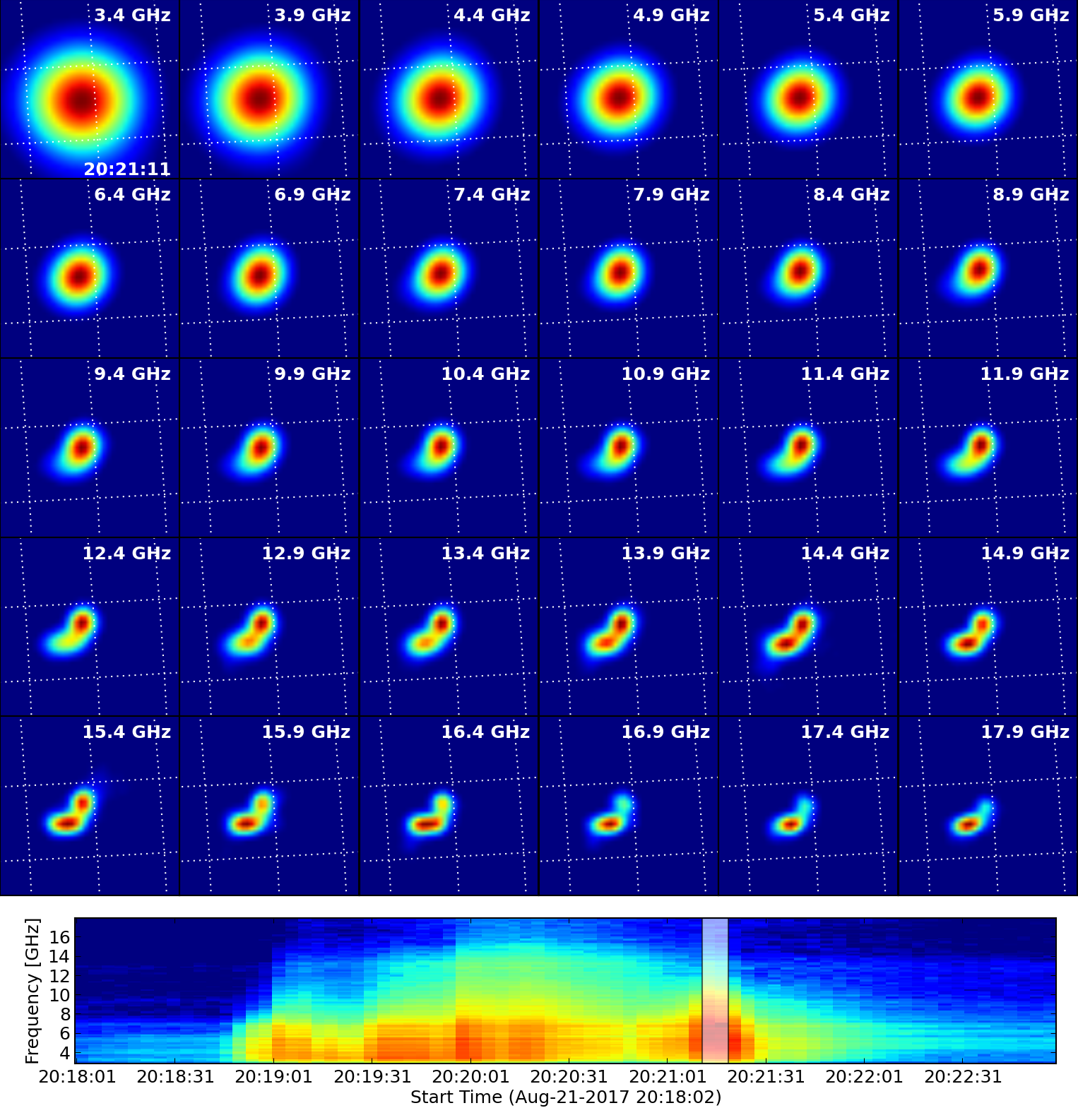
By default, qlookplot produces a full sun radio image (512x512 with a pixel size of 5"). If you know where the radio source is (e.g., from the previous full-Sun imaging), you can make a partial solar image around the source by specifying the image center ("xycen"), pixel scale ("cell"), and image field of view ("fov"). Here we show an example that images a 8-s interval around 20:21:14 UT using multi-frequency synthesis in 12-14 GHz and a smaller restoring beam. The microwave source is show to bifurcate into two components, which correspond pretty well with the double flare ribbons in SDO/AIA.
## in SunCASA
xycen = [375, 45] ## image center for clean in solar X-Y in arcsec
cell=['2.0arcsec'] ## pixel scale
imsize=[128] ## number of pixels in X and Y. If only one value is provided, NX = NY
fov = [100,100] ## field of view of the zoomed-in panels in unit of arcsec
ql.qlookplot(vis=msfile, specfile=specfile, timerange='20:21:10~20:21:18', \
spw='12GHz~14GHz', stokes='XX', restoringbeam=['6arcsec'],\
imsize=imsize,cell=cell,xycen=xycen,fov=fov,calpha=1.0)
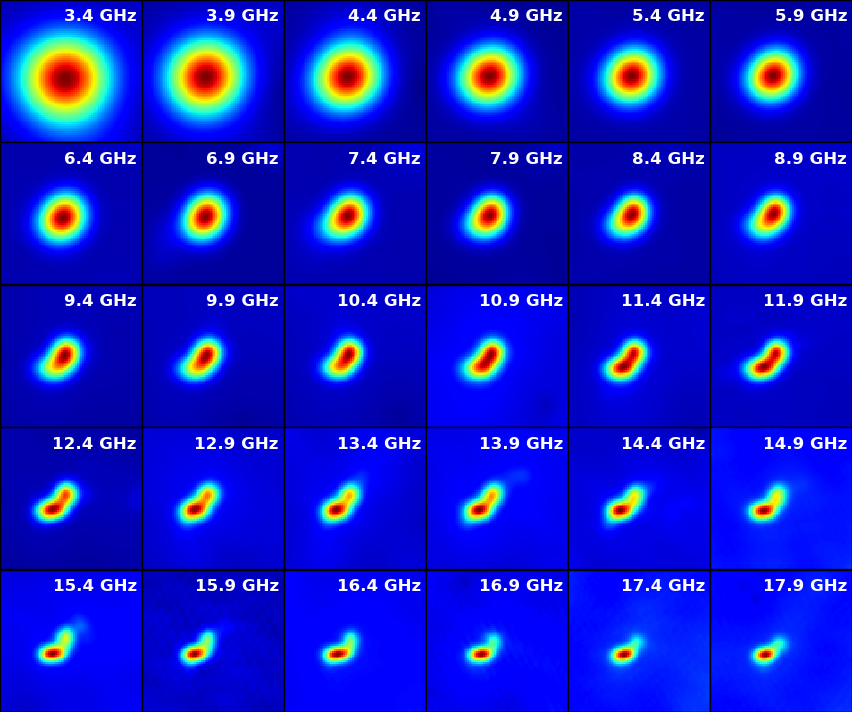
Next, we will make images for every single spectral window in this data set (from spw 1 to 30, spw 0 is in the visibility data, but is not calibrated for this event).
## in SunCASA
xycen = [375, 45] ## image center for clean in solar X-Y in arcsec
cell=['2.0arcsec'] ## pixel size
imsize=[128] ## x and y image size in pixels.
fov = [100,100] ## field of view of the zoomed-in panels in unit of arcsec
spw = ['{}'.format(s) for s in range(1,31)]
clevels = [0.5, 1.0] ## contour levels to fill in between.
calpha=0.35 ## now tune down the alpha
restoringbeam=['6arcsec']
ql.qlookplot(vis=msfile, specfile=specfile, timerange=timerange, spw=spw, stokes=stokes, \
restoringbeam=restoringbeam,imsize=imsize,cell=cell, \
xycen=xycen,fov=fov,clevels=clevels,calpha=calpha)
The output FITS file as a 4-d cube is saved to msfile + '.outim.image.fits' under your working directory. The 30 spectral windows used for spectral imaging are combined in the output FITS file. So NAXIS3 (frequency axis) is 30.
NAXIS = 4 / number of array dimensions NAXIS1 = 128 NAXIS2 = 128 NAXIS3 = 30 NAXIS4 = 2
Quick-Look Imaging series
# In SunCASA
msfile = 'IDB20170821201800-202300.4s.slfcaled.ms'
specfile = msfile + '.dspec.npz'
## set the time interval
timerange = '2017/08/21/20:21:10~2017/08/21/20:21:18'
## Bin width for time averaging
twidth = 1
## frequency range
spw = ['0~3']
## image center for clean in solar X-Y in arcsec
xycen = [375, 45]
## number of pixels in X and Y. If only one value is provided, NX = NY
imsize = [128]
## field of view of the zoomed-in panels in unit of arcsec
fov = [100., 100.]
## pixel scale
cell = ['2.0arcsec']
## select stokes XX
stokes = 'XX'
## for EOVSA data, set usemsphacenter to False
usemsphacenter = False
## set True if make a movie
mkmovie = True
## set True if generate compressed fits
docompress = True
#### ---- Control knobs for AIA plotting ---- ####
## set True if plot AIA images as the background
plotaia = True
## provide the path to the directory where the AIA fits files are located. Otherwise, set it to be ''
aiadir = 'Directory-where-AIA-fits-files-are-located'
## AIA passband. The options are [171,131,304,335,211,193,94,1600,1700]
aiawave = 171
## numbers of CPU threads for computing
ncpu = 2
ql.qlookplot(vis=msfile, specfile=specfile, timerange=timerange, spw=spw,
xycen=xycen, imsize=imsize, fov=fov, cell=cell, usemsphacenter=usemsphacenter,
plotaia=plotaia, aiadir=aiadir, aiawave=aiawave,
mkmovie=mkmovie, twidth=twidth, ncpu=ncpu, docompress=docompress, stokes=stokes)
Batch-Mode Imaging
This section is for interested users who wish to generate FITS files with full control on all parameters being used for synthesis imaging. We provide one example SunCASA script for generating 30-band spectral imaging maps, and another for iterating over time to produce a time series of these maps.
Producing a 30-band map cube for a given time
An example script can be found at this Github link. If you are on the AWS server Virgo, it is under /common/data/eovsa_tutorial/imaging_example.py. First, download or copy the script to your own working directory and cd to your directory.
# in SunCASA CASA <##>: cd your_working_directory CASA <##>: !cp /common/data/eovsa_tutorial/imaging_example.py ./
Second (optional), change inputs in the following block in your copy of the "imaging_example.py" script and save the changes. This block has definitions for time range, image center and FOV, antennas used, cell size, number of pixels, etc.
################### USER INPUT GOES IN THIS BLOK ################ vis = 'IDB20170821201800-202300.4s.slfcaled.ms' # input visibility data trange = '2017/08/21/20:21:00~2017/08/21/20:21:30' # time range for imaging xycen = [380., 50.] # center of the output map (in solar X and Y, Unit: arcsec) xran = [340., 420.] # plot range in solar X. Unit: arcsec yran = [10., 90.] # plot range in solar Y. Unit: arcsec antennas = '' # Use all 13 EOVSA 2-m antennas by default npix = 256 # number of pixels in the image cell = '1arcsec' # pixel scale in arcsec pol = 'XX' # polarization to image, use XX for now pbcor = True # correct for primary beam response? outdir = './images' # Specify where to save the output fits files outimgpre = 'EO' # Something to add to the output image name
Then, run the script in SunCASA via
CASA <##>: execfile('imaging_example.py')
The output is a combined 30-band FITS file saved under "outdir". The naming conversion is outimgpre + YYYYMMDDTHHMMSS_ALLBD.fits. In this example, it is "./images/EO20170821T202115.000_ALLBD.fits". Here is the detailed information of the axes of the output FITS file.
NAXIS = 4 / number of array dimensions NAXIS1 = 128 NAXIS2 = 128 NAXIS3 = 30 NAXIS4 = 2
From this point, you can use your favorite language to read the fits files and plot them. The last block of the example script uses SunPy.map to generate the plot shown on the right. For SSWIDL users, please refer to Section 3.3.3.3.
Producing a time series of 30-band maps (a 4-d cube)
An example script can be found at this Github link. If you are on the AWS server Virgo, it is under /common/data/eovsa_tutorial/imaging_timeseries_example.py. The script enables parallel-processing (based on a customized task "ptclean3" -- do not ask me why we have "3" in the task name). To invoke more CPU processes for parallel-processing, change "nthreads" in the preambles from 1 to the number of threads you wish to use.
Caution: This script is very computationally intensive. Please do not try to run this script on the AWS server during the RHESSI 18 live tutorial, as it has limited resources that everyone must share.
# in SunCASA CASA <##>: cd your_working_directory CASA <##>: !cp /common/data/eovsa_tutorial/imaging_timeseries_example.py ./
Similar as the previous script, change inputs in the following block of your copy of the "imaging_timeseries_example.py" script and save the changes.
################### USER INPUT GOES IN THIS BLOK #################### vis = 'IDB20170821201800-202300.4s.slfcaled.ms' # input visibility data specfile = vis + '.dspec.npz' ## input dynamic spectrum nthreads = 1 # Number of processing threads to use overwrite = True # whether to overwrite the existed fits files. trange = '' # time range for imaging, default to all times in the data twidth = 1 # make one image out of every 2 time integrations xycen = [380., 50.] # center of the output map in solar X and Y. Unit: arcsec xran = [340., 420.] # plot range in solar X. Unit: arcsec yran = [10., 90.] # plot range in solar Y. Unit: arcsec antennas = '' # default is to use all 13 EOVSA 2-m antennas. npix = 128 # number of pixels in the image cell = '2arcsec' # pixel scale in arcsec pol = 'XX' # polarization to image, use XX for now pbcor = True # correct for primary beam response? grid_spacing = 5. * u.deg # grid spacing in degrees outdir = './image_series/' # Specify where to save the output fits files imresfile = 'imres.npz' # File to write summary of the imaging results outimgpre = 'EO' # Something to add to the image name
Then, run the script in SunCASA by
CASA <##>: execfile('imaging_timeseries_example.py')
The output fits images and a summary file imresfile are saved under "outdir" (in this example "./image_timeseries"). The naming convention of output fits images is the same as the previous section. The summary yields the time, frequency, and path to every image.
CASA <##>: imres = np.load(outdir + imresfile)['imres'].item() CASA <##>: imres.keys() ['FreqGHz', 'ImageName', 'Succeeded', 'BeginTime', 'EndTime'] CASA <##>: ls image_series/*.fits | head -5 image_series/EO20170821T201800.500_ALLBD.fits image_series/EO20170821T201804.500_ALLBD.fits image_series/EO20170821T201808.500_ALLBD.fits image_series/EO20170821T201812.500_ALLBD.fits image_series/EO20170821T201816.500_ALLBD.fits
The last block of the example script uses SunPy.map to generate a series of plots and wraps them as a javascript movie.
Plotting Images in SSWIDL
All the output files are in standard FITS format in the Helioprojective Cartesian coordinate system (that most spacecraft solar image data adopt; FITS header CTYPE is HPLN-TAN and HPLT-TAN). They are fully compatible with the SSWIDL map suite which deals with FITS files. We have prepared SSWIDL routines to convert the single time or time-series FITS files to an array of SSWIDL map structure. The scripts are available in the Github repository of our tutorial. For those working on Virgo, local copies are placed under /common/data/eovsa_tutorial/. Two scripts are relevant to this tutorial:
- casa_readfits.pro: read an array of multi-frequency FITS files into FITS header (index) and data.
- casa_fits2map.pro: convert an array of multi-frequency FITS files into an array of SSWIDL map structure (adapted from P. T. Gallagher's hsi_fits2map.pro)
First, start SSWIDL from command line:
sswidl
Find and convert the fits files:
; cd to your path that contains casa_readfits.pro and casa_fits2map.pro. If on Virgo, just add the path IDL> add_path, '/common/data/eovsa_tutorial/' IDL> fitsdir = '/common/data/eovsa_tutorial/image_series/' IDL> fitsfiles = file_search(fitsdir+'*_ALLBD.fits') ; The resulting fitfiles contains all multi-frequency FITS files under "fitsdir" ; The following command converts the fits files to an array of map structures. ; "casa_fits2map.pro" also work well on a single FITS file. ; (Optional) keyword "calcrms" is to calculate rms and dynamic range (SNR) of the maps in a user-defined "empty" region ; of the map specified by "rmsxran" and "rmsyran" ;;; Here we load 10 time frames as a demonstration IDL> casa_fits2map, fitsfiles[45:54], eomaps [, /calcrms, rmsxran=[260., 320.], rmsyran=[-70., 0.]]
The resulting maps have a shape of [# of frequencies, # of polarizations, # of times]
IDL> help,eomaps EOMAPS STRUCT = -> <Anonymous> Array[30, 1, 10]
They have compatible keywords recognized by, e.g., plot_map.pro. Example of the output map keywords:
IDL> help,eomaps,/str ** Structure <36009f8>, 24 tags, length=131336, data length=131332, refs=1: DATA DOUBLE Array[128, 128] XC DOUBLE 378.99175 YC DOUBLE 49.001472 DX DOUBLE 2.0000000 DY DOUBLE 2.0000000 TIME STRING '21-Aug-2017 20:20:59.500' ID STRING 'EOVSA XX 3.413GHz' DUR DOUBLE 3.9999997 XUNITS STRING 'arcsec' YUNITS STRING 'arcsec' ROLL_ANGLE DOUBLE 0.0000000 ROLL_CENTER DOUBLE Array[2] FREQ DOUBLE 3.4125694 FREQUNIT STRING 'GHz' STOKES STRING 'XX' DATAUNIT STRING 'K' DATATYPE STRING 'Brightness Temperature' BMAJ DOUBLE 0.0097377778 BMIN DOUBLE 0.0097377778 BPA DOUBLE 0.0000000 RSUN DOUBLE 948.03989 L0 FLOAT 0.00000 B0 DOUBLE 6.9299225 COMMENT STRING 'Converted by CASA_FITS2MAP.PRO'
An example for plotting all images at a selected time:
; choose a time pixel
IDL> plttime = anytim('2017-08-21T20:21:15')
IDL> dt = min(abs(anytim(eomaps[0,0,*].time)-plttime),tidx)
; use maps at this time, first polarization, and all bands
IDL> eomap = reform(eomaps[*,0,tidx])
IDL> window,0,xs=1000,ys=800
IDL> !p.multi=[0,6,5]
IDL> loadct, 3
IDL> for i=0, 29 do plot_map,eomap[i],grid=5.,$
tit=string(eomap[i].freq,format='(f5.2)')+' GHz', $
chars=2.0,xran=[340.,420.],yran=[10.,90.]
Spectral Fitting with GSFIT
In this section we discuss using GSFIT (now a package of SSWIDL) to perform spectral fit based on the resulting FITS images produced from the previous steps. We will use the FITS images produced in Section 3.3.3.2 as the input. On Virgo, we prepared an IDL save file located at /common/data/eovsa_tutorial/21August2017.sav. In addition, this folder contains three more demo files that may be used to demonstrate the GSFIT functionality. For those intending to use their own machines for this part of the tutorial, these IDL fits files may be downloaded from this location
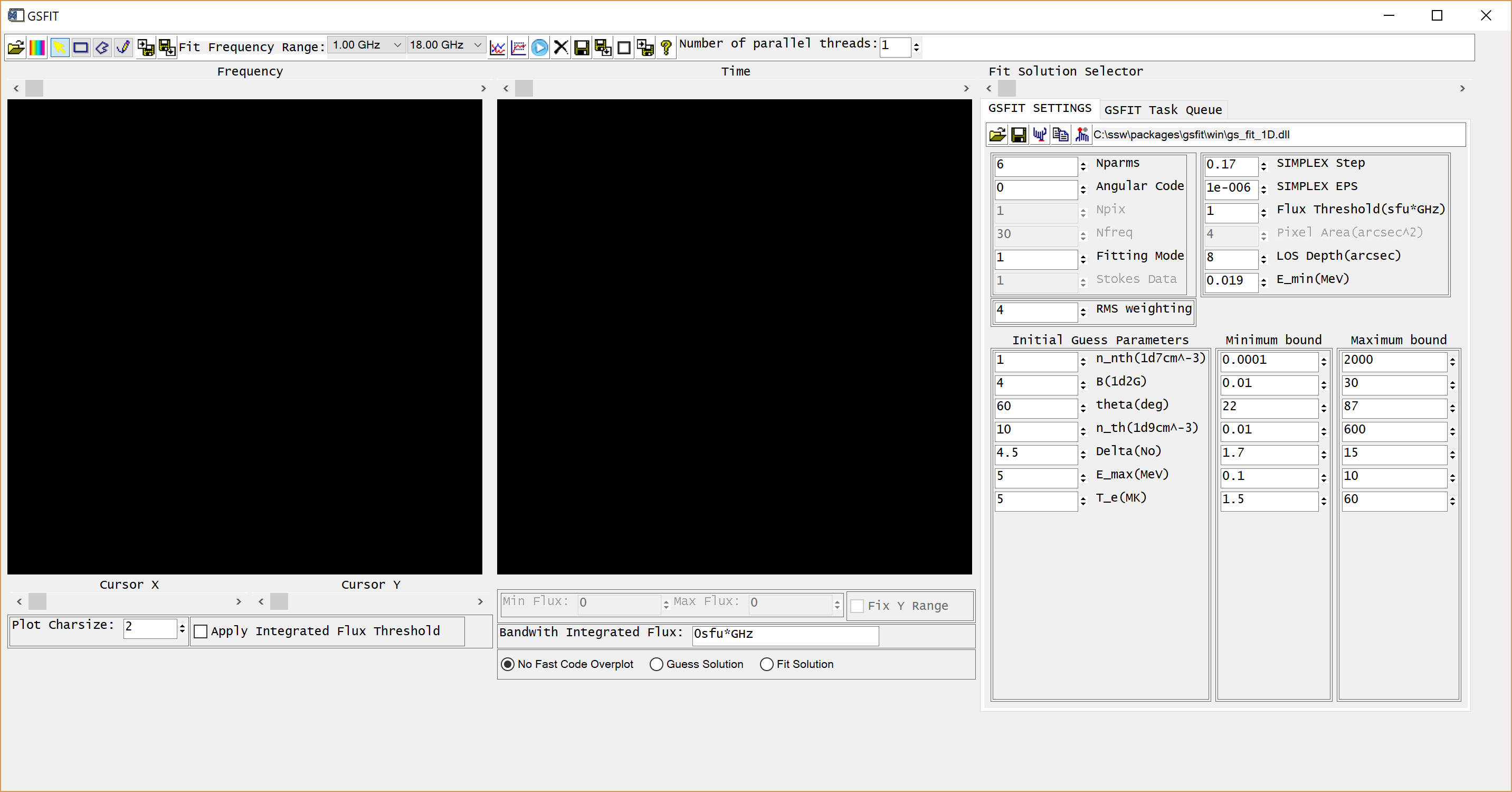
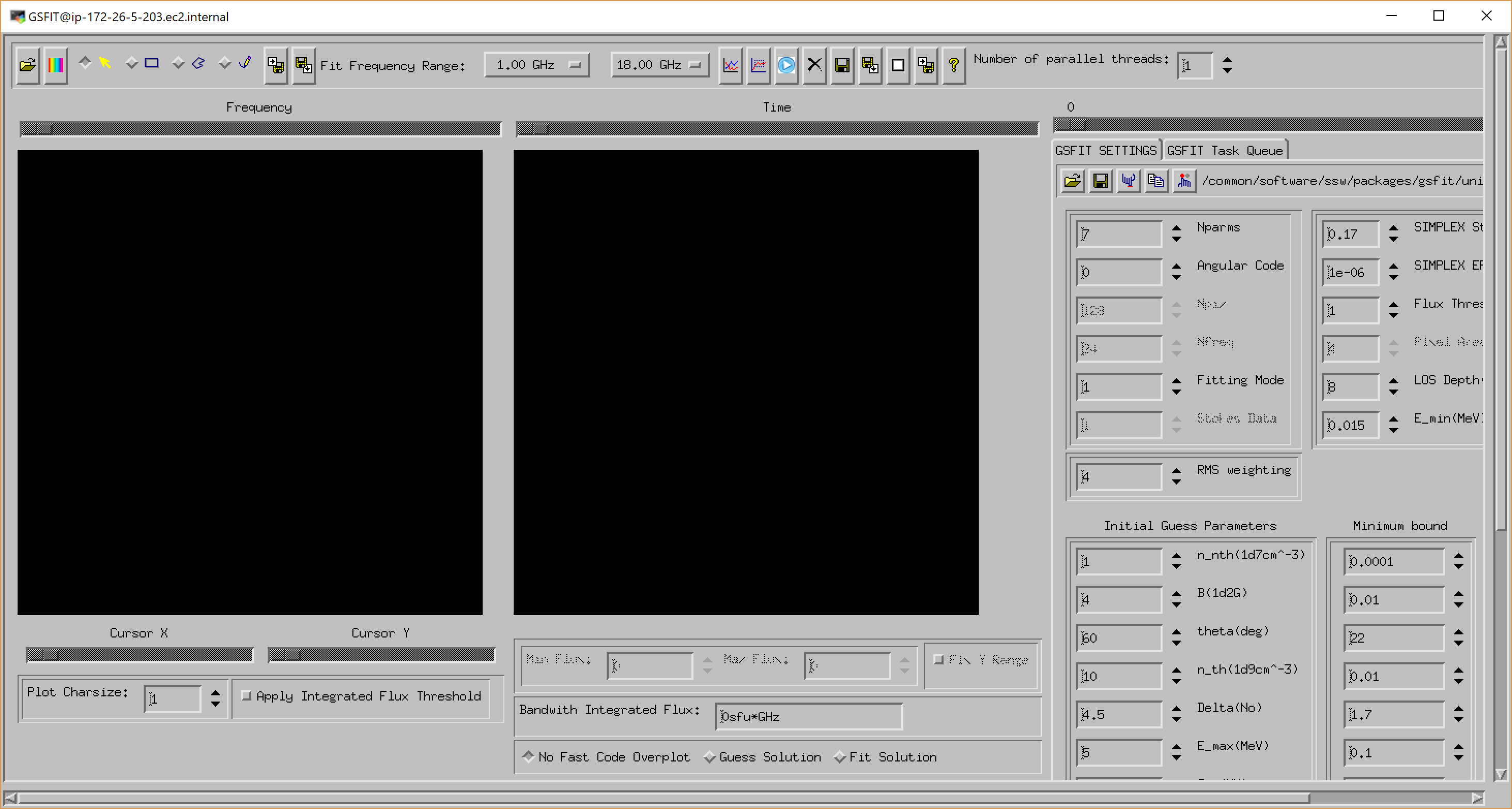
GSFIT GUI Application
The GSFIT GUI application may be launched as follows
IDL> gsfit [,nthreads]
where nthreads is an optional argument that indicates the number of parallel asynchronous threads to be used when performing the fit tasks. By default, GSFIT launches with only one thread, but the user may interactively add or delete threads as needed at the run-time up to the number of CPUs available on the system (Note, if you are working on the AWS server, please use only one thread). After some delay while the interface loads, the GUI below should appear.
A detailed description of the GSFIT functionality is provided in the page linked below.
GSFIT Help
GSFITCP Batch Mode Application
GSFITCP is the command prompt counterpart of the GSFIT GUI microwave spectral fitting application. Once started, GSFITCP is designed to run in unattended mode until all fitting tasks assigned to it are completed and log on the disk in an user-defined *.log file, the content of which may be visualized using the GSFITVIEW GUI application. When run remotely on an Linux/Mac platform, the GSFITCP may be launched on a detached screen, which allows the remote user to logout without stopping the process in which GSFITCP runs.
GSFITCP is launched using the following call:
IDL> gsfitcp, taskfile, nthreads, /start
where taskfile is a path to a file in which a GSFIT task has been previously saved, as explained in the GSFIT Help page, nthreads is an optional argument indicating the number of parallel asynchronous threads to be used, and the optional keyword /start, if set, requests immediate start of the batch processing.
A detailed description of the GSFITCP functionality is provided in the page linked below.
GSFITCP Help
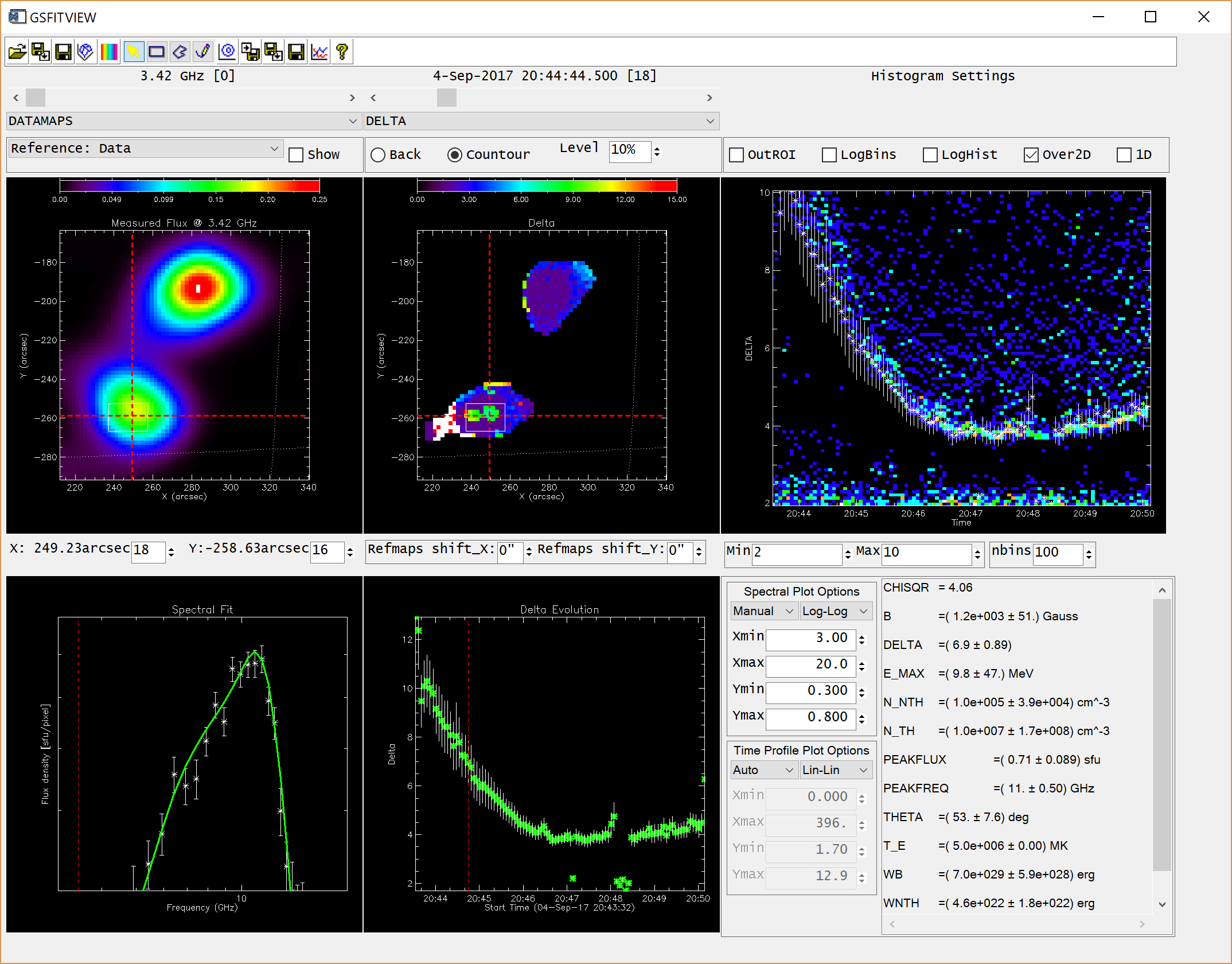
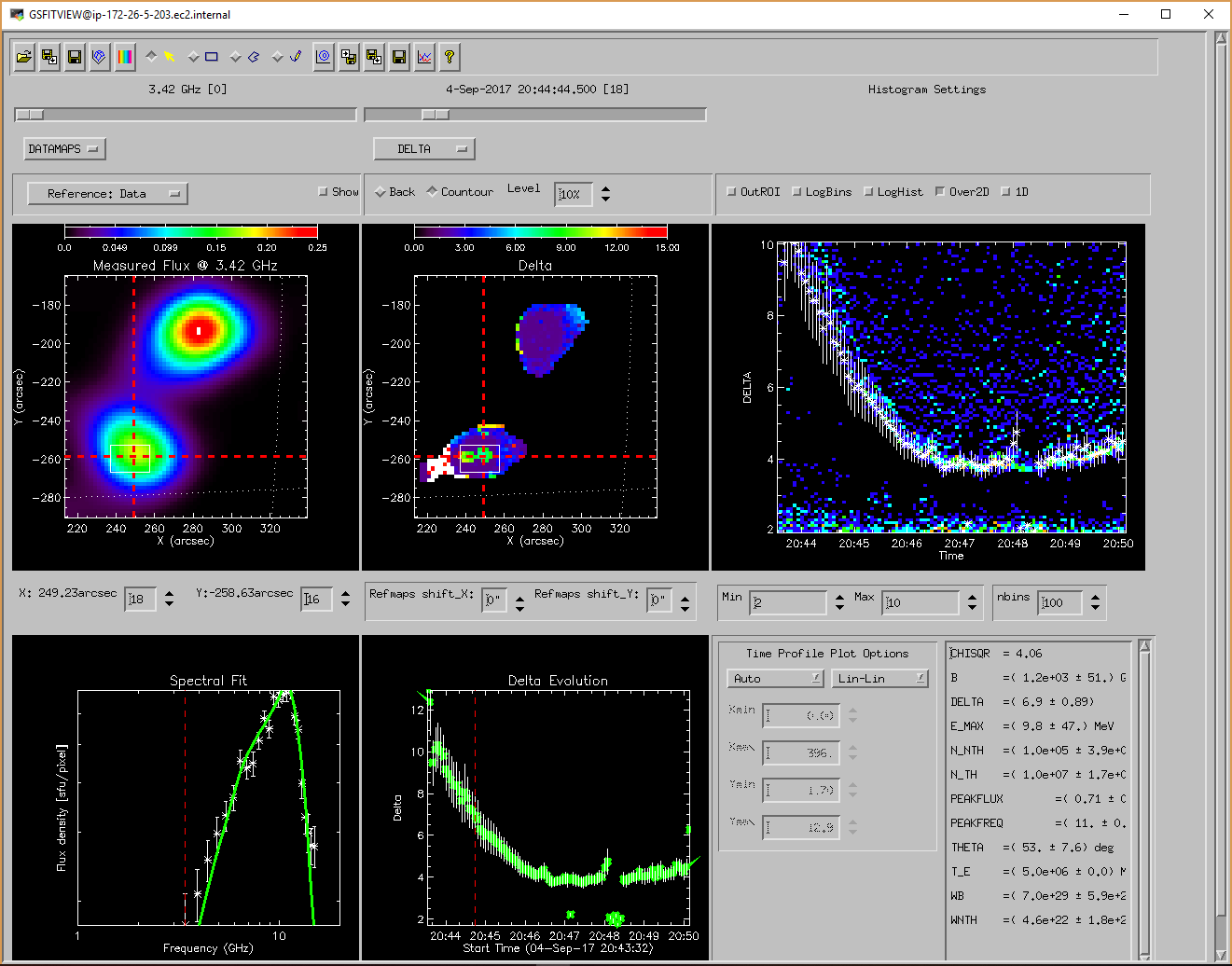
GSFITVIEW GUI Application
The GSFITVIEW GUI application may be launched as follows
IDL> gsfitview [,gsfitmaps]
where the optional gsfitmaps argument is either the filename of an IDL *.sav file containg a GSFIT Parameter Map Cube structure produced by the GSFIT or GSFITCP applications, or an already restored such structure.
A detailed description of the GSFITVIEW functionality is provided in the page linked below.